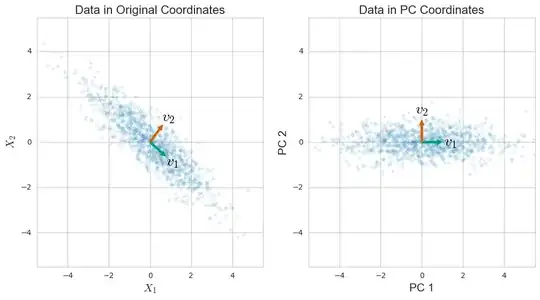
Let me start with PCA. Suppose that you have n data points comprised of d numbers (or dimensions) each. If you center this data (subtract the mean data point $\mu$ from each data vector $x_i$) you can stack the data to make a matrix
$$
X = \left(
\begin{array}{ccccc}
&& x_1^T - \mu^T && \\
\hline
&& x_2^T - \mu^T && \\
\hline
&& \vdots && \\
\hline
&& x_n^T - \mu^T &&
\end{array}
\right)\,.
$$
The covariance matrix
$$
S = \frac{1}{n-1} \sum_{i=1}^n (x_i-\mu)(x_i-\mu)^T = \frac{1}{n-1} X^T X
$$
measures to which degree the different coordinates in which your data is given vary together. So, it's maybe not surprising that PCA -- which is designed to capture the variation of your data -- can be given in terms of the covariance matrix. In particular, the eigenvalue decomposition of $S$ turns out to be
$$
S = V \Lambda V^T = \sum_{i = 1}^r \lambda_i v_i v_i^T \,,
$$
where $v_i$ is the $i$-th Principal Component, or PC, and $\lambda_i$ is the $i$-th eigenvalue of $S$ and is also equal to the variance of the data along the $i$-th PC. This decomposition comes from a general theorem in linear algebra, and some work does have to be done to motivate the relatino to PCA.
SVD is a general way to understand a matrix in terms of its column-space and row-space. (It's a way to rewrite any matrix in terms of other matrices with an intuitive relation to the row and column space.) For example, for the matrix $A = \left( \begin{array}{cc}1&2\\0&1\end{array} \right)$ we can find directions $u_i$ and $v_i$ in the domain and range so that
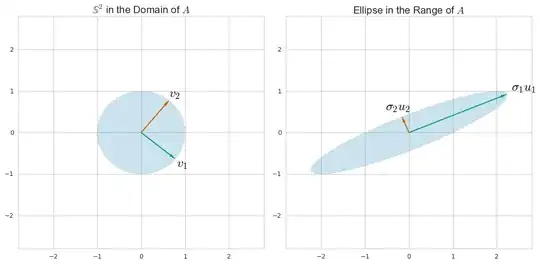
You can find these by considering how $A$ as a linear transformation morphs a unit sphere $\mathbb S$ in its domain to an ellipse: the principal semi-axes of the ellipse align with the $u_i$ and the $v_i$ are their preimages.
In any case, for the data matrix $X$ above (really, just set $A = X$), SVD lets us write
$$
X = \sum_{i=1}^r \sigma_i u_i v_j^T\,,
$$
where $\{ u_i \}$ and $\{ v_i \}$ are orthonormal sets of vectors.A comparison with the eigenvalue decomposition of $S$ reveals that the "right singular vectors" $v_i$ are equal to the PCs, the "right singular vectors" are
$$
u_i = \frac{1}{\sqrt{(n-1)\lambda_i}} Xv_i\,,
$$
and the "singular values" $\sigma_i$ are related to the data matrix via
$$
\sigma_i^2 = (n-1) \lambda_i\,.
$$
It's a general fact that the right singular vectors $u_i$ span the column space of $X$. In this specific case, $u_i$ give us a scaled projection of the data $X$ onto the direction of the $i$-th principal component. The left singular vectors $v_i$ in general span the row space of $X$, which gives us a set of orthonormal vectors that spans the data much like PCs.
I go into some more details and benefits of the relationship between PCA and SVD in this longer article.