I found something intresting today!
I have always been doing PCA by using SVD with covariance when I want to reduce the dimension.
Assume that we have a vector $X = {x_0, x_1, x_2, \dots, x_n}$. $X$ contains lots of noise, but it's actually a sine curve. Let's remove that noise.
Covariance method:
X = X - mean(X) % Center the dataX = cov(X'*X); % Find covariance[U, S, V] = svd(X, 'econ'); % Use Singular value decompotisionnx = 1; % DimensionX = U(:, 1:nx)*S(1:nx, 1:nx)*V(:, 1:nx)'; % Dimension reduce
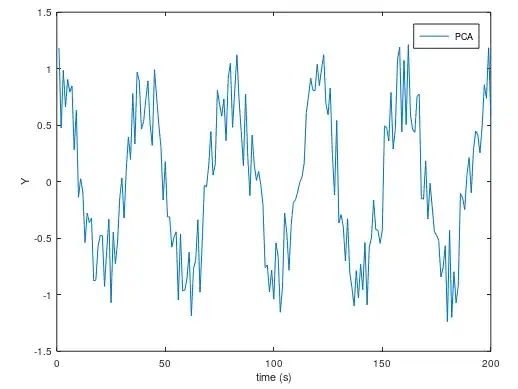
When I plot $X$, I got this. The amplitude is better (it should be 1), but the noise is still there.
So I tried this instead.
n = size(X, 2);H = hankel(X);H = H(1:n/2, 1:n/2); % Important with half hankelnx = 1; % Dimension[U, S, V] = svd(H, 'econ'); % Use Singular value decompotisionX = U(:, 1:nx)*S(1:nx, 1:nx)*V(:, 1:nx)'; % Dimension reduce
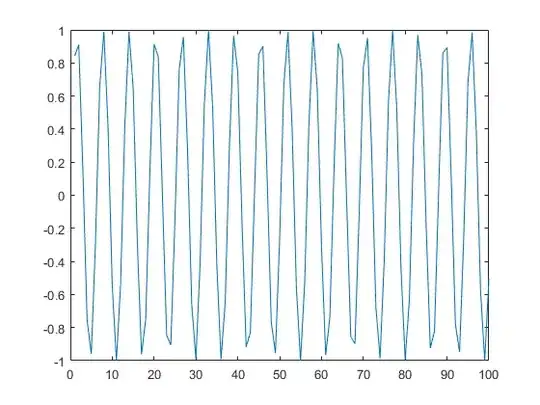
And now I got this result. A much better result.
Question:
Is it better to use a hankel matrix instead of covariance matrix when reducing dimensions of data using PCA?