I am trying to enhance the contrast in the images I get after scanning a surface using Thermography (Principal Component Thermography ~Rajic, which is basically an application of Principal Component Analysis) to see the defects clearly.
But my question here is to understand the difference between empirical orthogonal functions and Principal Components as shown below: 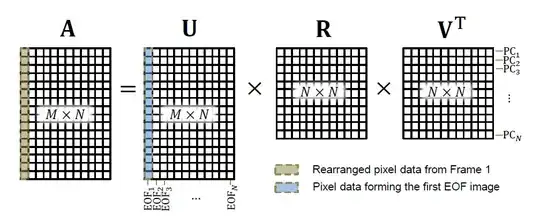 SVD is used to decompose matrix
SVD is used to decompose matrix A into U, R, and V.
In my case, I have 20 images. And after doing a raster like operation to unroll an image in a column, I get my matrix A which has 20 columns and each of them represents one image.
Any intuitive explanation will be really appreciated.