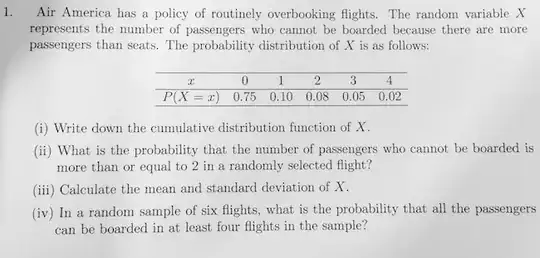
I have some data which looks like this when I plot a normalized histogram.
The full data set is available here and here (the second link is pastebin). It is 20,000 lines long.
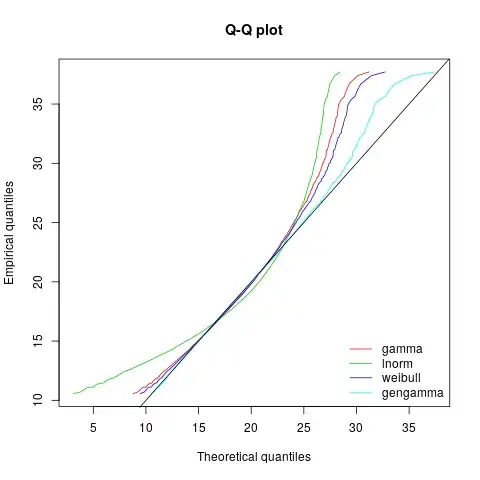
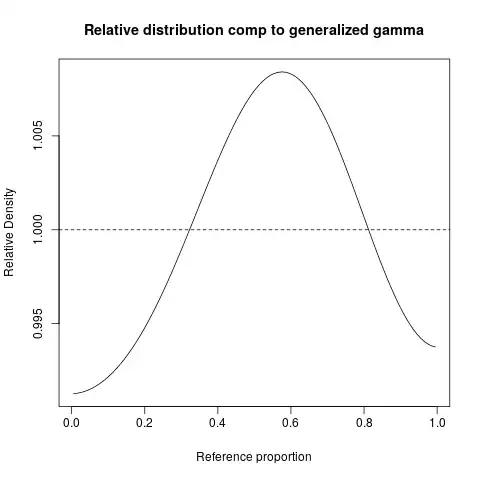
My guess is that it is a sample from a (generalized) gamma distribution but I have failed to show this.
I attempted in python to fit a generalized gamma distribution using
stats.gengamma.fit(data)
but it returns
(12.406504335648364, 0.89229023296508592, 9.0627571605857646, 0.51700107895010139)
and I am not sure what to make of it.
Overall, how can I work out what distribution my data is likely to be and how could I test it in R or preferably python?
Assuming my coding/understanding is not broken, it now seems unlikely this data is from a generalised gamma distribution.
- I simulated 100 samples of size 20,000 using the parameters (12.406504335648364, 0.89229023296508592, 9.0627571605857646, 0.51700107895010139) given above.
- I computed the Kolmogorov-Smirnov statistic for each using
stats.kstest(simul_data, 'gengamma', args = (a,c,loc,scale)).statistic. - I found that the Kolmogorov-Smirnov statistic for the data is larger than all 100 from the simulation.