Maybe a bit late, but I would like to add an answer here for future knowledge.
One way to find the best $\epsilon$ for DBSCAN is to compute the knn, then sort the distances and see where the "knee" is located.
Example in python, because is the language I manage.:
from sklearn.neighbors import NearestNeighbors
import plotly.express as px
neighbors = 6
# X_embedded is your data
nbrs = NearestNeighbors(n_neighbors=neighbors ).fit(X_embedded)
distances, indices = nbrs.kneighbors(X_embedded)
distance_desc = sorted(distances[:,ns-1], reverse=True)
px.line(x=list(range(1,len(distance_desc )+1)),y= distance_desc )
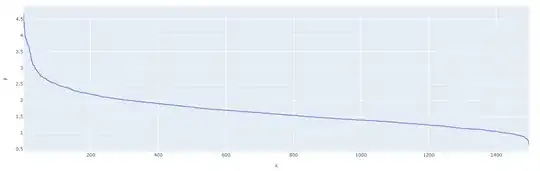
Then, to find the "knee", you can use another package:
(pip install kneed)
from kneed import KneeLocator
kneedle = KneeLocator(range(1,len(distanceDec)+1), #x values
distanceDec, # y values
S=1.0, #parameter suggested from paper
curve="convex", #parameter from figure
direction="decreasing") #parameter from figure
To see where the "knee" is, you can run
kneedle.plot_knee_normalized()
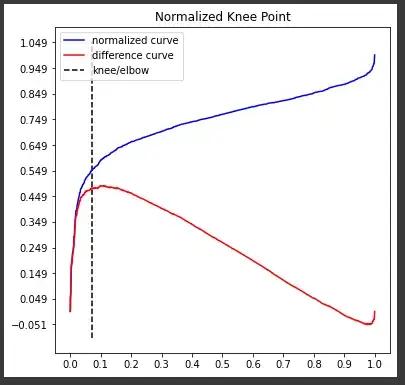
the commands kneedle.elbow or kneedle.knee returns the index of the x array, and the kneedle.knee_y returns the optimum value for $\epsilon$.

