I am starting to dabble with the use of glmnet with LASSO Regression where my outcome of interest is dichotomous. I have created a small mock data frame below:
age <- c(4, 8, 7, 12, 6, 9, 10, 14, 7)
gender <- c(1, 0, 1, 1, 1, 0, 1, 0, 0)
bmi_p <- c(0.86, 0.45, 0.99, 0.84, 0.85, 0.67, 0.91, 0.29, 0.88)
m_edu <- c(0, 1, 1, 2, 2, 3, 2, 0, 1)
p_edu <- c(0, 2, 2, 2, 2, 3, 2, 0, 0)
f_color <- c("blue", "blue", "yellow", "red", "red", "yellow", "yellow",
"red", "yellow")
asthma <- c(1, 1, 0, 1, 0, 0, 0, 1, 1)
# df is a data frame for further use!
df <- data.frame(age, gender, bmi_p, m_edu, p_edu, f_color, asthma)
The columns (variables) in the above dataset are as follows:
age(age of child in years) - continuousgender- binary (1 = male; 0 = female)bmi_p(BMI percentile) - continuousm_edu(mother highest education level) - ordinal (0 = less than high school; 1 = high school diploma; 2 = bachelors degree; 3 = post-baccalaureate degree)p_edu(father highest education level) - ordinal (same as m_edu)f_color(favorite primary color) - nominal ("blue", "red", or "yellow")asthma(child asthma status) - binary (1 = asthma; 0 = no asthma)
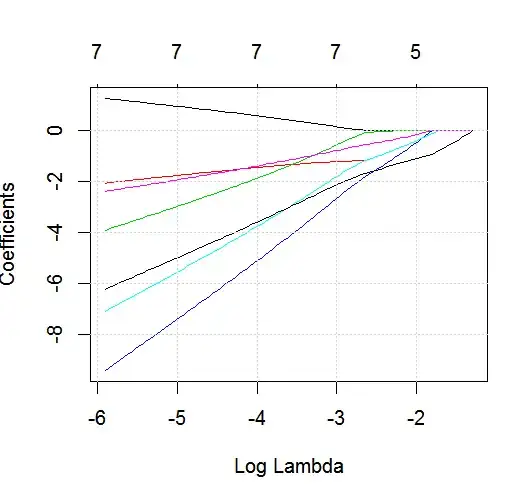
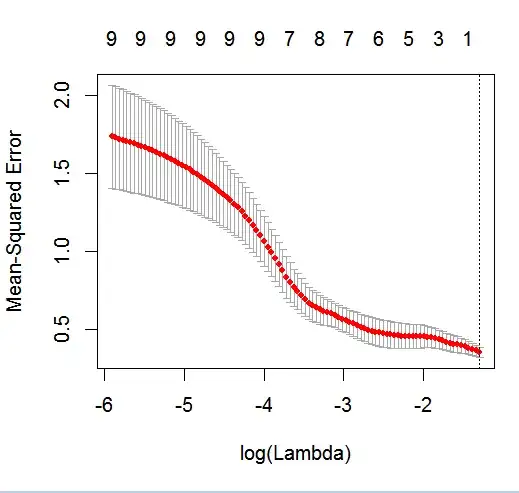
The goal of this example is to make use of LASSO to create a model predicting child asthma status from the list of 6 potential predictor variables (age, gender, bmi_p, m_edu, p_edu, and f_color). Obviously the sample size is an issue here, but I am hoping to gain more insight into how to handle the different types of variables (i.e., continuous, ordinal, nominal, and binary) within the glmnet framework when the outcome is binary (1 = asthma; 0 = no asthma).
As such, would anyone being willing to provide a sample R script along with explanations for this mock example using LASSO with the above data to predict asthma status? Although very basic, I know I, and likely many others on CV, would greatly appreciate this!