In general, one of the easiest ways to sample from a mixture distribution is the following:
Algorithm Steps
1) Generate a random variable $U\sim\text{Uniform}(0,1)$
2) If $U\in\left[\sum_{i=1}^kp_{k},\sum_{i=1}^{k+1}p_{k+1}\right)$ interval, where $p_{k}$ correspond to the the probability of the $k^{th}$ component of the mixture model, then generate from thedistribution of the $k^{th}$ component
3) Repeat steps 1) and 2) until you have the desired amount of samples from the mixture distribution
Now using the general algorithm given above, you could sample from your example mixture of normals by using the following R code:
#The number of samples from the mixture distribution
N = 100000
#Sample N random uniforms U
U =runif(N)
#Variable to store the samples from the mixture distribution
rand.samples = rep(NA,N)
#Sampling from the mixture
for(i in 1:N){
if(U[i]<.3){
rand.samples[i] = rnorm(1,0,1)
}else if(U[i]<.8){
rand.samples[i] = rnorm(1,10,1)
}else{
rand.samples[i] = rnorm(1,3,.1)
}
}
#Density plot of the random samples
plot(density(rand.samples),main="Density Estimate of the Mixture Model")
#Plotting the true density as a sanity check
x = seq(-20,20,.1)
truth = .3*dnorm(x,0,1) + .5*dnorm(x,10,1) + .2*dnorm(x,3,.1)
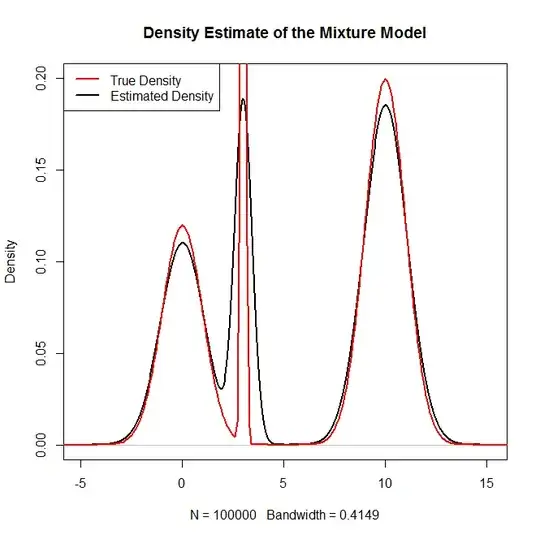
plot(density(rand.samples),main="Density Estimate of the Mixture Model",ylim=c(0,.2),lwd=2)
lines(x,truth,col="red",lwd=2)
legend("topleft",c("True Density","Estimated Density"),col=c("red","black"),lwd=2)
Which generates:
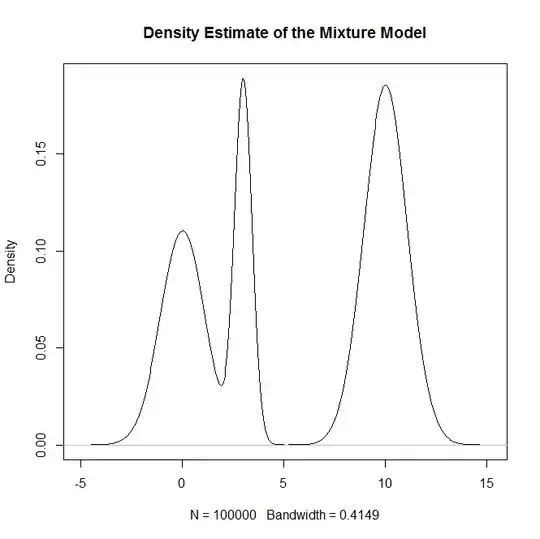
and as a sanity check: