As I understand your problem, the main issue is the size of the data set, and not that it contains missing value (i.e. "sparse"). For such a problem, I would recommend doing a partial PCA in order to solve for a subset of leading PCs. The package irlba allows for this by performing a "Lanczos bidiagonalization". It is much faster for large matrices when you are only interested in returning a few of the leading PCs. In the following example, I have adapted a bootstrapping technique that I discussed here into a function that incorporates this method as well as a variable sub-sampling parameter. In the function bootpca, you can define the number of variables to sample, n, the number of PCs to return, npc, and the number of iterations B for the sub-sampling routine. For this method, I have centered and scaled the sub-sampled matrix in order to standardize the variance of the dataset and allow for comparability among the singular values of the matrix decomposition. By making a boxplot of these bootstrapped singular values, lam, you should be able to differentiate between PCs that carry signals from those that are dominated by noise.
Example
Generate data
m=50
n=100
x <- (seq(m)*2*pi)/m
t <- (seq(n)*2*pi)/n
#field
Xt <-
outer(sin(x), sin(t)) +
outer(sin(2.1*x), sin(2.1*t)) +
outer(sin(3.1*x), sin(3.1*t)) +
outer(tanh(x), cos(t)) +
outer(tanh(2*x), cos(2.1*t)) +
outer(tanh(4*x), cos(0.1*t)) +
outer(tanh(2.4*x), cos(1.1*t)) +
tanh(outer(x, t, FUN="+")) +
tanh(outer(x, 2*t, FUN="+"))
Xt <- t(Xt)
image(Xt)
#Noisy field
set.seed(1)
RAND <- matrix(runif(length(Xt), min=-1, max=1), nrow=nrow(Xt), ncol=ncol(Xt))
R <- RAND * 0.2 * Xt
#True field + Noise field
Xp <- Xt + R
image(Xp)
load bootpca function
library(irlba)
bootpca <- function(mat, n=0.5*nrow(mat), npc=10, B=40*nrow(mat)){
lam <- matrix(NaN, nrow=npc, ncol=B)
for(b in seq(B)){
samp.b <- NaN*seq(n)
for(i in seq(n)){
samp.b[i] <- sample(nrow(mat), 1)
}
mat.b <- scale(mat[samp.b,], center=TRUE, scale=TRUE)
E.b <- irlba(mat.b, nu=npc, nv=npc)
lam[,b] <- E.b$d
print(paste(round(b/B*100), "%", " completed", sep=""))
}
lam
}
Result and plot
res <- bootpca(Xp, n=0.5*nrow(Xp), npc=15, B=999) #50% of variables used in each iteration, 15 PCs computed, and 999 iterations
par(mar=c(4,4,1,1))
boxplot(t(res), log="y", col=8, outpch="", ylab="Lambda [log-scale]")
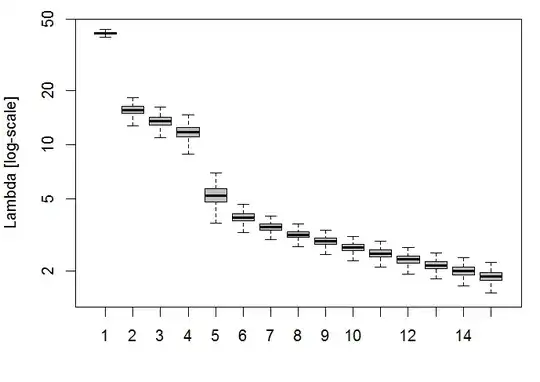
It's obvious that the leading 5 PCs carry the most information, although there were technically 9 signals in the example data set.
For your very large data set, you may want to use a smaller fraction of variables (i.e. rows) in each iteration, but do many iterations.
