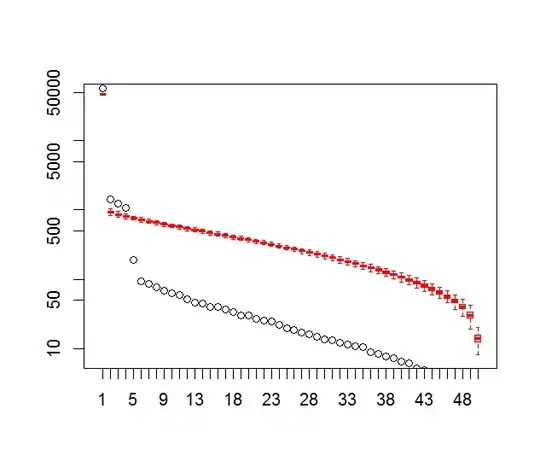
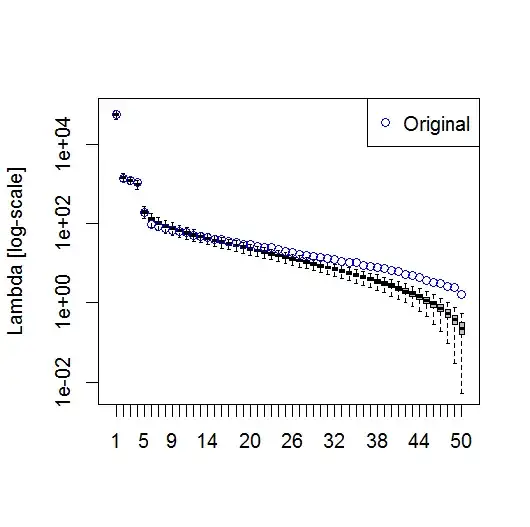
I am interested in determining the number of significant patterns coming out of a Principal Component Analysis (PCA) or Empirical Orthogonal Function (EOF) Analysis. I am particularly interested in applying this method to climate data. The data field is a MxN matrix with M being the time dimension (e.g. days) and N being the spatial dimension (e.g. lon/lat locations). I have read of a possible bootstrap method to determine significant PCs, but have been unable to find a more detailed description. Until now, I have been applying North's Rule of Thumb (North et al., 1982) to determine this cutoff, but I was wondering if a more robust method was available.
As an example:
###Generate data
x <- -10:10
y <- -10:10
grd <- expand.grid(x=x, y=y)
#3 spatial patterns
sp1 <- grd$x^3+grd$y^2
tmp1 <- matrix(sp1, length(x), length(y))
image(x,y,tmp1)
sp2 <- grd$x^2+grd$y^2
tmp2 <- matrix(sp2, length(x), length(y))
image(x,y,tmp2)
sp3 <- 10*grd$y
tmp3 <- matrix(sp3, length(x), length(y))
image(x,y,tmp3)
#3 respective temporal patterns
T <- 1:1000
tp1 <- scale(sin(seq(0,5*pi,,length(T))))
plot(tp1, t="l")
tp2 <- scale(sin(seq(0,3*pi,,length(T))) + cos(seq(1,6*pi,,length(T))))
plot(tp2, t="l")
tp3 <- scale(sin(seq(0,pi,,length(T))) - 0.2*cos(seq(1,10*pi,,length(T))))
plot(tp3, t="l")
#make data field - time series for each spatial grid (spatial pattern multiplied by temporal pattern plus error)
set.seed(1)
F <- as.matrix(tp1) %*% t(as.matrix(sp1)) +
as.matrix(tp2) %*% t(as.matrix(sp2)) +
as.matrix(tp3) %*% t(as.matrix(sp3)) +
matrix(rnorm(length(T)*dim(grd)[1], mean=0, sd=200), nrow=length(T), ncol=dim(grd)[1]) # error term
dim(F)
image(F)
###Empirical Orthogonal Function (EOF) Analysis
#scale field
Fsc <- scale(F, center=TRUE, scale=FALSE)
#make covariance matrix
C <- cov(Fsc)
image(C)
#Eigen decomposition
E <- eigen(C)
#EOFs (U) and associated Lambda (L)
U <- E$vectors
L <- E$values
#projection of data onto EOFs (U) to derive principle components (A)
A <- Fsc %*% U
dim(U)
dim(A)
#plot of top 10 Lambda
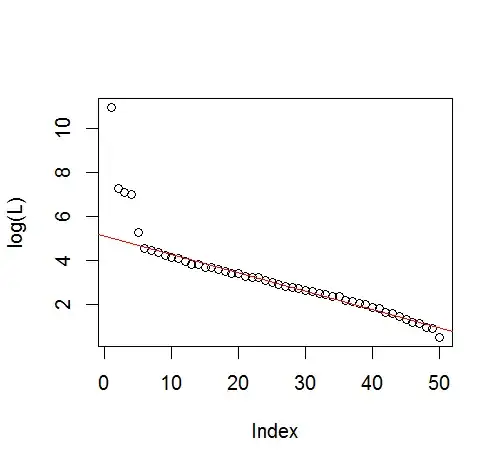
plot(L[1:10], log="y")
#plot of explained variance (explvar, %) by each EOF
explvar <- L/sum(L) * 100
plot(explvar[1:20], log="y")
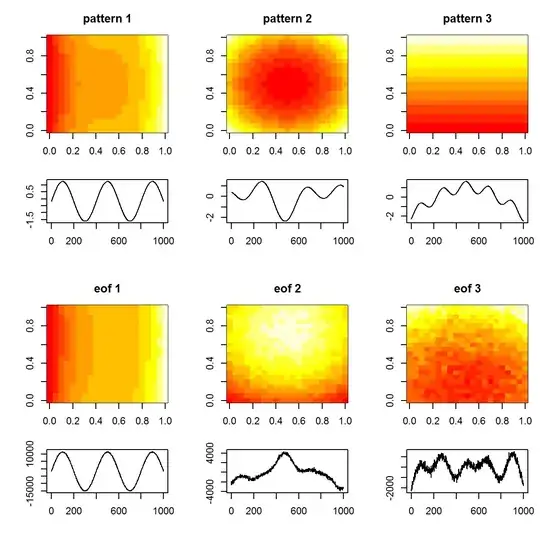
#plot original patterns versus those identified by EOF
layout(matrix(1:12, nrow=4, ncol=3, byrow=TRUE), widths=c(1,1,1), heights=c(1,0.5,1,0.5))
layout.show(12)
par(mar=c(4,4,3,1))
image(tmp1, main="pattern 1")
image(tmp2, main="pattern 2")
image(tmp3, main="pattern 3")
par(mar=c(4,4,0,1))
plot(T, tp1, t="l", xlab="", ylab="")
plot(T, tp2, t="l", xlab="", ylab="")
plot(T, tp3, t="l", xlab="", ylab="")
par(mar=c(4,4,3,1))
image(matrix(U[,1], length(x), length(y)), main="eof 1")
image(matrix(U[,2], length(x), length(y)), main="eof 2")
image(matrix(U[,3], length(x), length(y)), main="eof 3")
par(mar=c(4,4,0,1))
plot(T, A[,1], t="l", xlab="", ylab="")
plot(T, A[,2], t="l", xlab="", ylab="")
plot(T, A[,3], t="l", xlab="", ylab="")
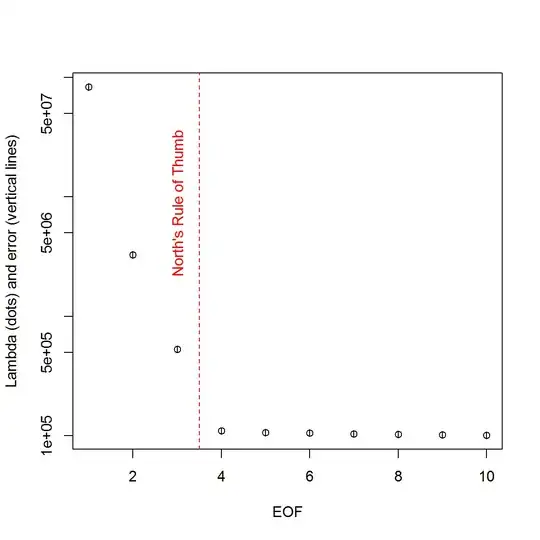
And, here is the method that I have been using to determine PC significance. Basically, the rule of thumb is that the difference between neighboring Lambdas must be greater than their associated error.
###Determine significant EOFs
#North's Rule of Thumb
Lambda_err <- sqrt(2/dim(F)[2])*L
upper.lim <- L+Lambda_err
lower.lim <- L-Lambda_err
NORTHok=0*L
for(i in seq(L)){
Lambdas <- L
Lambdas[i] <- NaN
nearest <- which.min(abs(L[i]-Lambdas))
if(nearest > i){
if(lower.lim[i] > upper.lim[nearest]) NORTHok[i] <- 1
}
if(nearest < i){
if(upper.lim[i] < lower.lim[nearest]) NORTHok[i] <- 1
}
}
n_sig <- min(which(NORTHok==0))-1
plot(L[1:10],log="y", ylab="Lambda (dots) and error (vertical lines)", xlab="EOF")
segments(x0=seq(L), y0=L-Lambda_err, x1=seq(L), y1=L+Lambda_err)
abline(v=n_sig+0.5, col=2, lty=2)
text(x=n_sig, y=mean(L[1:10]), labels="North's Rule of Thumb", srt=90, col=2)
I have found the chapter section by Björnsson and Venegas (1997) on significance tests to be helpful - they refer to three categories of tests, of which the dominant variance-type is probably what I am hoping to use. The refer to a type of Monte Carlo approach of shuffling the time dimension and recomputing the Lambdas over many permutations. von Storch and Zweiers (1999) also refer to the a test that compares the Lambda spectrum to a reference "noise" spectrum. In both cases, I am a bit unsure of how this might be done, and also how the significance test is done given the confidence intervals identified by the permutations.
Thanks for your help.
References: Björnsson, H. and Venegas, S.A. (1997). "A manual for EOF and SVD analyses of climate data", McGill University, CCGCR Report No. 97-1, Montréal, Québec, 52pp. http://andvari.vedur.is/%7Efolk/halldor/PICKUP/eof.pdf
G.R. North, T.L. Bell, R.F. Cahalan, and F.J. Moeng. (1982). Sampling errors in the estimation of empirical orthogonal functions. Mon. Wea. Rev., 110:699–706.
von Storch, H, Zwiers, F.W. (1999). Statistical analysis in climate research. Cambridge University Press.