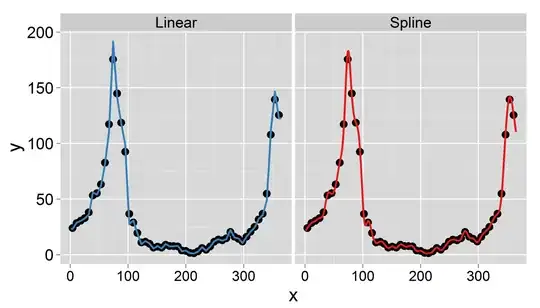
Edit
I have found a paper describing exactly the procedure I need. The only difference is that the paper interpolates monthly mean data to daily, while preserving the monthly means. I have trouble to implement the approach in R. Any hints are appreciated.
Original
For each week, I have the following count data (one value per week):
- Number of doctors' consultations
- Number of cases of influenza
My goal is to obtain daily data by interpolation (I thought of linear or truncated splines). The important thing is that I want to conserve the weekly mean, i.e. the mean of the daily interpolated data should equal the recorded value of this week. In addition, the interpolation should be smooth. One problem that could arise is that a certain week has less than 7 days (e.g. at the beginning or end of a year).
I would be grateful for advice on this matter.
Thanks a lot.
Here's a sample data set for the year 1995 (updated):
structure(list(daily.ts = structure(c(9131, 9132, 9133, 9134,
9135, 9136, 9137, 9138, 9139, 9140, 9141, 9142, 9143, 9144, 9145,
9146, 9147, 9148, 9149, 9150, 9151, 9152, 9153, 9154, 9155, 9156,
9157, 9158, 9159, 9160, 9161, 9162, 9163, 9164, 9165, 9166, 9167,
9168, 9169, 9170, 9171, 9172, 9173, 9174, 9175, 9176, 9177, 9178,
9179, 9180, 9181, 9182, 9183, 9184, 9185, 9186, 9187, 9188, 9189,
9190, 9191, 9192, 9193, 9194, 9195, 9196, 9197, 9198, 9199, 9200,
9201, 9202, 9203, 9204, 9205, 9206, 9207, 9208, 9209, 9210, 9211,
9212, 9213, 9214, 9215, 9216, 9217, 9218, 9219, 9220, 9221, 9222,
9223, 9224, 9225, 9226, 9227, 9228, 9229, 9230, 9231, 9232, 9233,
9234, 9235, 9236, 9237, 9238, 9239, 9240, 9241, 9242, 9243, 9244,
9245, 9246, 9247, 9248, 9249, 9250, 9251, 9252, 9253, 9254, 9255,
9256, 9257, 9258, 9259, 9260, 9261, 9262, 9263, 9264, 9265, 9266,
9267, 9268, 9269, 9270, 9271, 9272, 9273, 9274, 9275, 9276, 9277,
9278, 9279, 9280, 9281, 9282, 9283, 9284, 9285, 9286, 9287, 9288,
9289, 9290, 9291, 9292, 9293, 9294, 9295, 9296, 9297, 9298, 9299,
9300, 9301, 9302, 9303, 9304, 9305, 9306, 9307, 9308, 9309, 9310,
9311, 9312, 9313, 9314, 9315, 9316, 9317, 9318, 9319, 9320, 9321,
9322, 9323, 9324, 9325, 9326, 9327, 9328, 9329, 9330, 9331, 9332,
9333, 9334, 9335, 9336, 9337, 9338, 9339, 9340, 9341, 9342, 9343,
9344, 9345, 9346, 9347, 9348, 9349, 9350, 9351, 9352, 9353, 9354,
9355, 9356, 9357, 9358, 9359, 9360, 9361, 9362, 9363, 9364, 9365,
9366, 9367, 9368, 9369, 9370, 9371, 9372, 9373, 9374, 9375, 9376,
9377, 9378, 9379, 9380, 9381, 9382, 9383, 9384, 9385, 9386, 9387,
9388, 9389, 9390, 9391, 9392, 9393, 9394, 9395, 9396, 9397, 9398,
9399, 9400, 9401, 9402, 9403, 9404, 9405, 9406, 9407, 9408, 9409,
9410, 9411, 9412, 9413, 9414, 9415, 9416, 9417, 9418, 9419, 9420,
9421, 9422, 9423, 9424, 9425, 9426, 9427, 9428, 9429, 9430, 9431,
9432, 9433, 9434, 9435, 9436, 9437, 9438, 9439, 9440, 9441, 9442,
9443, 9444, 9445, 9446, 9447, 9448, 9449, 9450, 9451, 9452, 9453,
9454, 9455, 9456, 9457, 9458, 9459, 9460, 9461, 9462, 9463, 9464,
9465, 9466, 9467, 9468, 9469, 9470, 9471, 9472, 9473, 9474, 9475,
9476, 9477, 9478, 9479, 9480, 9481, 9482, 9483, 9484, 9485, 9486,
9487, 9488, 9489, 9490, 9491, 9492, 9493, 9494, 9495), class = "Date"),
wdayno = c(0L, 1L, 2L, 3L, 4L, 5L, 6L, 0L, 1L, 2L, 3L, 4L,
5L, 6L, 0L, 1L, 2L, 3L, 4L, 5L, 6L, 0L, 1L, 2L, 3L, 4L, 5L,
6L, 0L, 1L, 2L, 3L, 4L, 5L, 6L, 0L, 1L, 2L, 3L, 4L, 5L, 6L,
0L, 1L, 2L, 3L, 4L, 5L, 6L, 0L, 1L, 2L, 3L, 4L, 5L, 6L, 0L,
1L, 2L, 3L, 4L, 5L, 6L, 0L, 1L, 2L, 3L, 4L, 5L, 6L, 0L, 1L,
2L, 3L, 4L, 5L, 6L, 0L, 1L, 2L, 3L, 4L, 5L, 6L, 0L, 1L, 2L,
3L, 4L, 5L, 6L, 0L, 1L, 2L, 3L, 4L, 5L, 6L, 0L, 1L, 2L, 3L,
4L, 5L, 6L, 0L, 1L, 2L, 3L, 4L, 5L, 6L, 0L, 1L, 2L, 3L, 4L,
5L, 6L, 0L, 1L, 2L, 3L, 4L, 5L, 6L, 0L, 1L, 2L, 3L, 4L, 5L,
6L, 0L, 1L, 2L, 3L, 4L, 5L, 6L, 0L, 1L, 2L, 3L, 4L, 5L, 6L,
0L, 1L, 2L, 3L, 4L, 5L, 6L, 0L, 1L, 2L, 3L, 4L, 5L, 6L, 0L,
1L, 2L, 3L, 4L, 5L, 6L, 0L, 1L, 2L, 3L, 4L, 5L, 6L, 0L, 1L,
2L, 3L, 4L, 5L, 6L, 0L, 1L, 2L, 3L, 4L, 5L, 6L, 0L, 1L, 2L,
3L, 4L, 5L, 6L, 0L, 1L, 2L, 3L, 4L, 5L, 6L, 0L, 1L, 2L, 3L,
4L, 5L, 6L, 0L, 1L, 2L, 3L, 4L, 5L, 6L, 0L, 1L, 2L, 3L, 4L,
5L, 6L, 0L, 1L, 2L, 3L, 4L, 5L, 6L, 0L, 1L, 2L, 3L, 4L, 5L,
6L, 0L, 1L, 2L, 3L, 4L, 5L, 6L, 0L, 1L, 2L, 3L, 4L, 5L, 6L,
0L, 1L, 2L, 3L, 4L, 5L, 6L, 0L, 1L, 2L, 3L, 4L, 5L, 6L, 0L,
1L, 2L, 3L, 4L, 5L, 6L, 0L, 1L, 2L, 3L, 4L, 5L, 6L, 0L, 1L,
2L, 3L, 4L, 5L, 6L, 0L, 1L, 2L, 3L, 4L, 5L, 6L, 0L, 1L, 2L,
3L, 4L, 5L, 6L, 0L, 1L, 2L, 3L, 4L, 5L, 6L, 0L, 1L, 2L, 3L,
4L, 5L, 6L, 0L, 1L, 2L, 3L, 4L, 5L, 6L, 0L, 1L, 2L, 3L, 4L,
5L, 6L, 0L, 1L, 2L, 3L, 4L, 5L, 6L, 0L, 1L, 2L, 3L, 4L, 5L,
6L, 0L, 1L, 2L, 3L, 4L, 5L, 6L, 0L, 1L, 2L, 3L, 4L, 5L, 6L,
0L, 1L, 2L, 3L, 4L, 5L, 6L, 0L), month = c(1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2,
2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 3, 3, 3,
3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3,
3, 3, 3, 3, 3, 3, 3, 3, 3, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4,
4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4,
4, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5,
5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 6, 6, 6, 6, 6, 6,
6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6,
6, 6, 6, 6, 6, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7,
7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 8, 8,
8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8,
8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 9, 9, 9, 9, 9, 9, 9, 9, 9,
9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9,
9, 9, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10,
10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10,
10, 10, 10, 11, 11, 11, 11, 11, 11, 11, 11, 11, 11, 11, 11,
11, 11, 11, 11, 11, 11, 11, 11, 11, 11, 11, 11, 11, 11, 11,
11, 11, 11, 12, 12, 12, 12, 12, 12, 12, 12, 12, 12, 12, 12,
12, 12, 12, 12, 12, 12, 12, 12, 12, 12, 12, 12, 12, 12, 12,
12, 12, 12, 12), year = c(1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995,
1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995, 1995), yearday = 0:364,
no.influ.cases = c(NA, NA, NA, 168L, NA, NA, NA, NA, NA,
NA, 199L, NA, NA, NA, NA, NA, NA, 214L, NA, NA, NA, NA, NA,
NA, 230L, NA, NA, NA, NA, NA, NA, 267L, NA, NA, NA, NA, NA,
NA, 373L, NA, NA, NA, NA, NA, NA, 387L, NA, NA, NA, NA, NA,
NA, 443L, NA, NA, NA, NA, NA, NA, 579L, NA, NA, NA, NA, NA,
NA, 821L, NA, NA, NA, NA, NA, NA, 1229L, NA, NA, NA, NA,
NA, NA, 1014L, NA, NA, NA, NA, NA, NA, 831L, NA, NA, NA,
NA, NA, NA, 648L, NA, NA, NA, NA, NA, NA, 257L, NA, NA, NA,
NA, NA, NA, 203L, NA, NA, NA, NA, NA, NA, 137L, NA, NA, NA,
NA, NA, NA, 78L, NA, NA, NA, NA, NA, NA, 82L, NA, NA, NA,
NA, NA, NA, 69L, NA, NA, NA, NA, NA, NA, 45L, NA, NA, NA,
NA, NA, NA, 51L, NA, NA, NA, NA, NA, NA, 45L, NA, NA, NA,
NA, NA, NA, 63L, NA, NA, NA, NA, NA, NA, 55L, NA, NA, NA,
NA, NA, NA, 54L, NA, NA, NA, NA, NA, NA, 52L, NA, NA, NA,
NA, NA, NA, 27L, NA, NA, NA, NA, NA, NA, 24L, NA, NA, NA,
NA, NA, NA, 12L, NA, NA, NA, NA, NA, NA, 10L, NA, NA, NA,
NA, NA, NA, 22L, NA, NA, NA, NA, NA, NA, 42L, NA, NA, NA,
NA, NA, NA, 32L, NA, NA, NA, NA, NA, NA, 52L, NA, NA, NA,
NA, NA, NA, 82L, NA, NA, NA, NA, NA, NA, 95L, NA, NA, NA,
NA, NA, NA, 91L, NA, NA, NA, NA, NA, NA, 104L, NA, NA, NA,
NA, NA, NA, 143L, NA, NA, NA, NA, NA, NA, 114L, NA, NA, NA,
NA, NA, NA, 100L, NA, NA, NA, NA, NA, NA, 83L, NA, NA, NA,
NA, NA, NA, 113L, NA, NA, NA, NA, NA, NA, 145L, NA, NA, NA,
NA, NA, NA, 175L, NA, NA, NA, NA, NA, NA, 222L, NA, NA, NA,
NA, NA, NA, 258L, NA, NA, NA, NA, NA, NA, 384L, NA, NA, NA,
NA, NA, NA, 755L, NA, NA, NA, NA, NA, NA, 976L, NA, NA, NA,
NA, NA, NA, 879L, NA, NA, NA, NA), no.consultations = c(NA,
NA, NA, 15093L, NA, NA, NA, NA, NA, NA, 20336L, NA, NA, NA,
NA, NA, NA, 20777L, NA, NA, NA, NA, NA, NA, 21108L, NA, NA,
NA, NA, NA, NA, 20967L, NA, NA, NA, NA, NA, NA, 20753L, NA,
NA, NA, NA, NA, NA, 18782L, NA, NA, NA, NA, NA, NA, 19778L,
NA, NA, NA, NA, NA, NA, 19223L, NA, NA, NA, NA, NA, NA, 21188L,
NA, NA, NA, NA, NA, NA, 22172L, NA, NA, NA, NA, NA, NA, 21965L,
NA, NA, NA, NA, NA, NA, 21768L, NA, NA, NA, NA, NA, NA, 21277L,
NA, NA, NA, NA, NA, NA, 16383L, NA, NA, NA, NA, NA, NA, 15337L,
NA, NA, NA, NA, NA, NA, 19179L, NA, NA, NA, NA, NA, NA, 18705L,
NA, NA, NA, NA, NA, NA, 19623L, NA, NA, NA, NA, NA, NA, 19363L,
NA, NA, NA, NA, NA, NA, 16257L, NA, NA, NA, NA, NA, NA, 19219L,
NA, NA, NA, NA, NA, NA, 17048L, NA, NA, NA, NA, NA, NA, 19231L,
NA, NA, NA, NA, NA, NA, 20023L, NA, NA, NA, NA, NA, NA, 19331L,
NA, NA, NA, NA, NA, NA, 18995L, NA, NA, NA, NA, NA, NA, 16571L,
NA, NA, NA, NA, NA, NA, 15010L, NA, NA, NA, NA, NA, NA, 13714L,
NA, NA, NA, NA, NA, NA, 10451L, NA, NA, NA, NA, NA, NA, 14216L,
NA, NA, NA, NA, NA, NA, 16800L, NA, NA, NA, NA, NA, NA, 18305L,
NA, NA, NA, NA, NA, NA, 18911L, NA, NA, NA, NA, NA, NA, 17812L,
NA, NA, NA, NA, NA, NA, 18665L, NA, NA, NA, NA, NA, NA, 18977L,
NA, NA, NA, NA, NA, NA, 19512L, NA, NA, NA, NA, NA, NA, 17424L,
NA, NA, NA, NA, NA, NA, 14464L, NA, NA, NA, NA, NA, NA, 16383L,
NA, NA, NA, NA, NA, NA, 19916L, NA, NA, NA, NA, NA, NA, 18255L,
NA, NA, NA, NA, NA, NA, 20113L, NA, NA, NA, NA, NA, NA, 20084L,
NA, NA, NA, NA, NA, NA, 20196L, NA, NA, NA, NA, NA, NA, 20184L,
NA, NA, NA, NA, NA, NA, 20261L, NA, NA, NA, NA, NA, NA, 22246L,
NA, NA, NA, NA, NA, NA, 23030L, NA, NA, NA, NA, NA, NA, 10487L,
NA, NA, NA, NA)), .Names = c("daily.ts", "wdayno", "month",
"year", "yearday", "no.influ.cases", "no.consultations"), row.names = c(NA,
-365L), class = "data.frame")