What I would like to do
I would like to reconstruct a logistic regression model with splines (Lymph Node Involvement (Cores)) using published coefficients and spline knots. All sources that I posted here are from: https://www.mskcc.org/nomograms/prostate/pre_op/coefficients
Why I would like to do that
My aim is to reconstruct the model "Lymph node involment (Cores)" in R, so that I can apply it on a large number of patients of a clinical study (without having to type all the data in an online calculator for every subject) and predict their probability of Lymph node involvement.
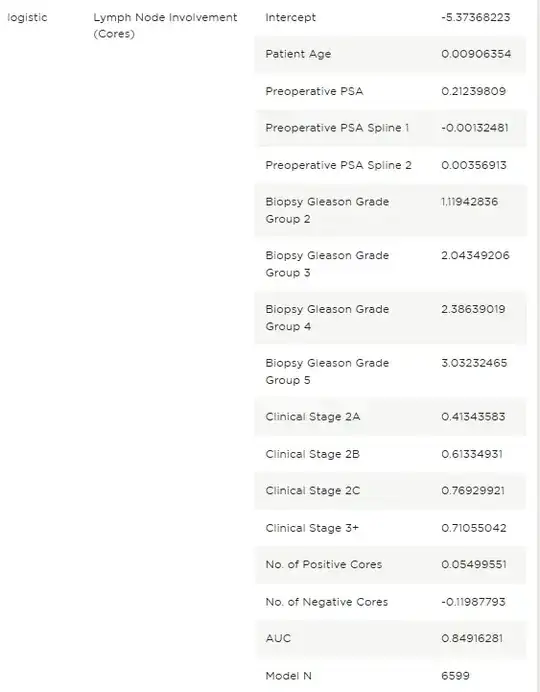
The published information about the model
The model definition is as follows:
The restricted cubic spline terms are as follows:
The intercept and coefficients are as follow:
Question
- Is it possible to reconstruct this model with the published data
- If yes, how can this be achieved using R
What I have already done
I found following source, however, it is slightly different (I do not want to change anything of the model) Reconstructing a logistic regression model from literature using published coefficients
I understand that I somehow have to reconstruct the model with something like:
#reconstruct model
copylogit <- ...
And then apply it to my data:
#make test data
newdata <-data.frame(age=as.numeric(80),psa=as.numeric(10),gleason_grade=as.factor(4),clinical_stage=as.character("2A"), no_of_positive_cores=as.numeric(2),no_of_negativ_cores=as.numeric(10))
#apply model to test data
predict(copylogit, newdata = newdata, type = "response")
And the expected result for this example would be: 17%
Update: Since I am not sure if it is possible to reconstruct the model with the published data I thought about to generate a prediction equation so that I can calculate the probabilites of lymph node involvement in a larger dataset. Therefore, I opened a new question: Calculate spline terms of a logistic regression using published knots and formula