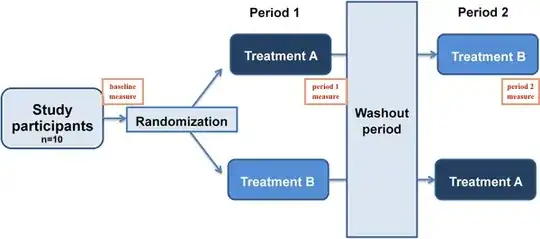
I'm working on a dataset of 2x2 crossover study. I have 10 subjects, each of them underwent both A and B treatment but in a different sequence. (This is a balanced study.)
I want to see how A and B treatments improve lipid levels. My thought process was to create a linear mixed model with subjects as a random effect; treatment, sequence, and period as the fixed effects; finally, sex and age as covariates.
My data:
#Reproducible data
id <- rep(1:10,3)
age <- rep(c("59","59","70","67","66","70","70","68","71","57"),3)
sex <- rep(c("F","M","F","M","F","F","F","M","F","M"),3)
sequence <- rep(c("1","2","1","2","1","2","1","2","2","2"),3)
period <- c(rep(0,10),rep(1,10),rep(2,10))
Treatment <- c(rep("C",10), rep(c("A","B"),4), "B","B",rep(c("B","A"),4), "A","A") #C is baseline
lipid <- c(18,6,30,12,14,19,10,22,22,27,13,28,14,23,12,27,9,10,13,22,13,22,29,12,16,24,15,13,17,11)
DF <- data.frame(id,age,sex,sequence,period,Treatment,lipid)
> head(DF)
id age sex sequence period Treatment lipid
1 1 59 F 1 0 C 18
2 2 59 M 2 0 C 6
3 3 70 F 1 0 C 30
4 4 67 M 2 0 C 12
5 5 66 F 1 0 C 14
6 6 70 F 2 0 C 19
My linear mixed model:
library(lmerTest)
lm1 <- lmer(lipid~Treatment + sequence + period + sex + age + (1|id), data = DF, REML = F)
> summary(lm1)
Random effects:
Groups Name Variance Std.Dev.
id (Intercept) 1.344 1.159
Residual 34.986 5.915
Number of obs: 30, groups: id, 10
Fixed effects:
Estimate Std. Error df t value Pr(>|t|)
(Intercept) 23.7890 18.2664 10.1410 1.302 0.222
TreatmentA -2.8500 2.8572 20.0000 -0.997 0.330
TreatmentB 2.2750 3.1018 20.0000 0.733 0.472
sequence2 4.7080 3.3324 10.0000 1.413 0.188
period1 -1.1250 2.6998 20.0000 -0.417 0.681
sexM -3.8351 3.7742 10.0000 -1.016 0.334
age -0.1078 0.2734 10.0000 -0.394 0.702
After building a linear mixed model, I wanted to do post-hoc test to compare treatment A and B. I tried both emmeans and multcomp but they are giving me different results.
Emmeans:
library(emmeans)
emm <- emmeans(lm1,"Treatment")
pairs(emm, adjust = "fdr")
> pairs(emm, adjust = "fdr")
contrast estimate SE df t.ratio p.value
C - A nonEst NA NA NA NA
C - B nonEst NA NA NA NA
A - B -5.12 2.93 23.5 -1.750 0.2794
Multicomp:
library(multcomp)
summary(glht(lm1, linfct = mcp(Treatment = "Tukey")), test = adjusted("fdr"))
> summary(glht(lm1, linfct = mcp(Treatment = "Tukey")), test = adjusted("fdr"))
Linear Hypotheses:
Estimate Std. Error z value Pr(>|z|)
A - C == 0 -2.850 2.857 -0.997 0.463
B - C == 0 2.275 3.102 0.733 0.463
B - A == 0 5.125 2.700 1.898 0.173
(Adjusted p values reported -- fdr method)
I guess question would be,
1) Based on the study design, does my linear mixed model lm1 <- lmer(lipid~Treatment + sequence + period + sex + age + (1|id), data = DF, REML = F) look ok? Or should I account for other interaction terms (ex. Treatment*sequence)?
2) Why does emmeans give me NAs in C-A and C-B when multcomp gives me values? Which one would you recommend to conduct the post-hoc test on lmer model since the results are different?
Any thought is appreciated, thank you!