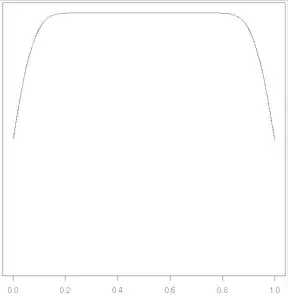
When using the density function in R, it includes smooth transitions down to 0 at both ends of the data. Is there a way to prevent this? As a trivial example, suppose I am calculating the density function for 1000 uniformly spaced points between 0 and 1. What I'd like is a straight line from 0 to 1. Instead, I get a straight line from about 0.2 to 0.8 with smooth transitions down to 0. (Numbers are a little different for non-default kernels but the same general idea.) Of course, I know the density in this example, but not in real examples. I can shrink these end caps using bw or adjust, but that causes an undesirable reduction in smoothing within the interval.
Is there an option to truncate the kernel at the boundaries of the region, so that it estimates the density using only points inside the region? I have worked around this by mirroring all of my points about the two ends and then keeping only the part in the middle, but that seems like a crazy hack for something that should be simple. Is there a simpler way to do this?