What sort of kernel density estimator does one use to avoid boundary bias?
Consider the task of estimating the density $f_0(x)$ with bounded support and where the probability mass is not decreasing or going to zero as the boundary is approached. To simplify matters assume that the bound(s) of the density is known.
To focus ideas consider as an example the uniform distribution:
Given a sample size $N$ of iid draws $\mathcal U(0,1)$ one could think of applying the kernel density estimator
$$\hat f(y) = \frac{1}{ns}\sum_i K\left( \frac{x_i-y}{s} \right)$$
with a normal kernel and some smoothing parameter $s$. To illustrate the boundary bias consider (implemented in the software R: A Language and Environment for Statistical Computing):
N <- 10000
x <- runif(N)
s <- .045
M <- 100
y <- seq(0,1,length.out=M)
out <- rep(0,M)
for (i in 1:M)
{
weights <- dnorm((x-y[i])/s)
out[i] <- mean(weights)/s
}
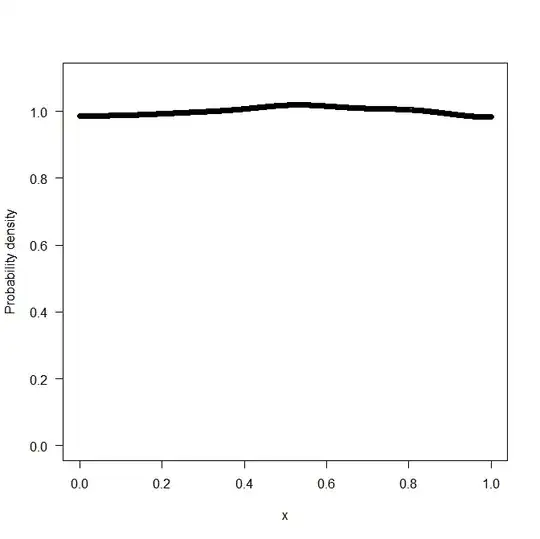
plot(y,out,type="l",ylim=c(0,1.5))
which generates the following plot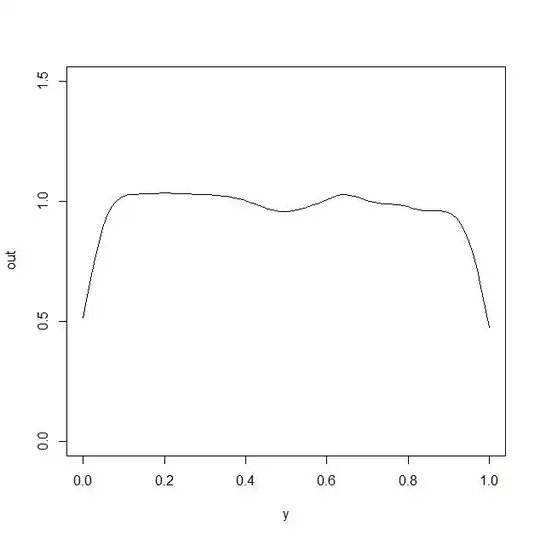
clearly the approach has a problem capturing the true value of the density function $f_0(x)$ at $x$ close to the boundary.
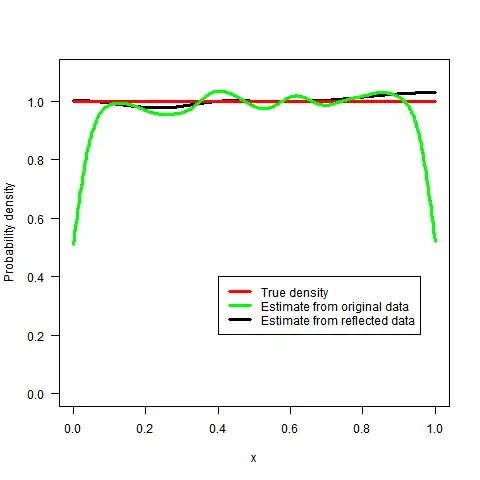
The logspline method works better but is certainly not without some boundary bias
library(logspline)
set.seed(1)
N <- 10000
x <- runif(N)
m <- logspline(x,lbound=0,ubound=1,knots=seq(0,1,length.out=21))
plot(m)