I have 214 covariates and a binary outcome. The total number of positive and negative outcomes is 27 and 33, respectively.
I already modelled it using a 1v1 univariable logistic regression and I found some significative coefficients.
After that I moved to a simple LASSO shrinkage and I obtained
> fit <- glmnet(x = fitXData, y = fitYData, family = "binomial", alpha = 1, nlambda = 50)
Call: glmnet(x = fitXData, y = fitYData, family = "binomial", alpha = 1, nlambda = 50)
Df %Dev Lambda
[1,] 0 8.067e-16 0.156700
[2,] 2 1.804e-02 0.142700
[3,] 3 3.901e-02 0.129900
[4,] 4 5.950e-02 0.118200
[5,] 5 7.772e-02 0.107600
....
Now let's assume that I want to use a model composed with the two most significative covariates provided by lasso and to compare it with the real outcome. Being a binary outcome I wrote
>table(fitYData,predict(fit, s=0.142700, newx= fitXData, type="class"))
fitYData FALSE
FALSE 33
TRUE 27
meaning that the prediction is FALSE for all the outcomes.
Chosing a different lambda (see later for the specific value) i obtain better results:
> table(fitYData,predict(fit, s=0.05574419, newx= fitXData, type="class"))
fitYData FALSE TRUE
FALSE 29 4
TRUE 10 17
Clearly it's due to the fact that the big "lambda" (0.1427) chosen to remove the 212 less significative covariates strongly impacts (lower) the "beta"s predictors resulting in a fully negative prediction.
It seems a very common behaviour to me for big lambda values (for instance, see https://www.rstatisticsblog.com/data-science-in-action/lasso-regression/, before the section Sharing the R Squared formula, all the values are underestimated) but nobody mentions it.
Point 1:
Should I perform and ordinary glm fit including only the two variables I selected or should I blindly trust on the glmnet prediction for this big lambda?
Point 2:
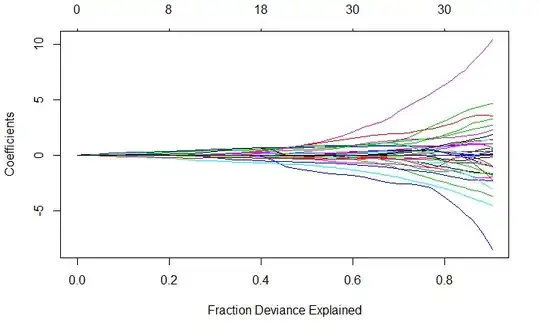
It seems to me that the correlation with the outcome is so small that it's very difficult to extract a good piece of information from it. I report the LASSO plot:
plot(fit, xvar = "dev")
The plot is not as clear as the ones I find in the online examples and papers... Is it normal?
Point 3:
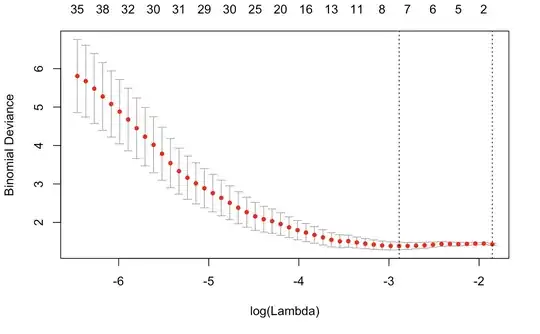
I also performed a CV Lasso with a leave-one-out k-fold:
cvfit <- cv.glmnet(fitXData, fitYData, family="binomial", grouped=FALSE, type.measure = "deviance", nfold = 60, nlambda = 50)
I report the CV results:
cvfit\$lambda.min = 0.05574419
cvfit\$lambda.1se = 0.1567398
With lambda.min 8 covariates has been "selected" but with lambda.1se all the variables are rejected.
Again, is it due to the low intrinsic correlation/high noise or am I mistaking? Shouldn't the deviance increase for big lambdas? The usual trend of this last graph is usually the opposite...
Thank you for your help!! :)
Best regards. Giulio