What are the assumptions when doing hypothesis testing using a Gamma GLM or GLMM? Are the residuals suppose to be normally distributed and is heteroscedasticity a concern like the Gaussian (normal) distribution? Do you test the assumptions in the same fashion Levene Tests of individual fixed effects, residual and qqplots, and Shapiro-Wilks test?
Discussion of when to use Gamma Glm and Glmm is found here:
However, I found little to no discussion of the assumptions and how to test them?
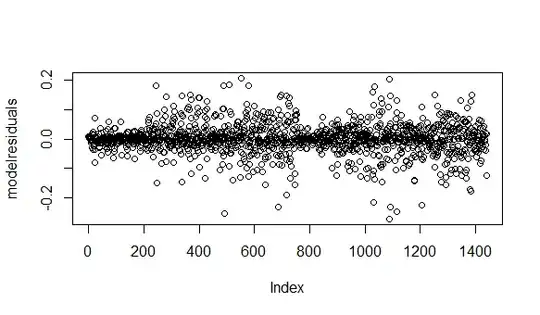
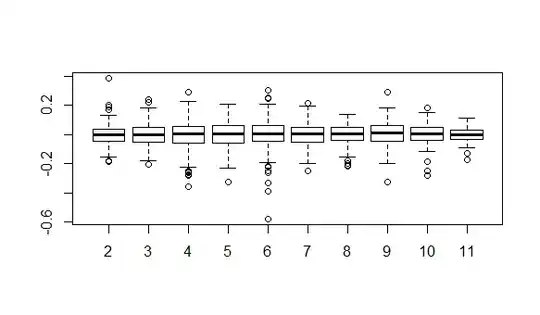
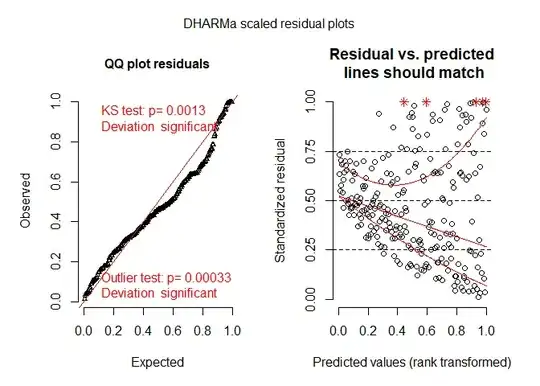
Edit 1: Requested Example residual plots- are any of these a concern. Levene's test, comes back significant.
Residuals are plotted using this code. Time (Obs) is the ordinal factor levels from the data
modelresiduals = residuals(Gama_model)
plot(modelresiduals)
plot(y = modelresiduals, x = Model_data2$Obs)
1. All resids-
2. By time
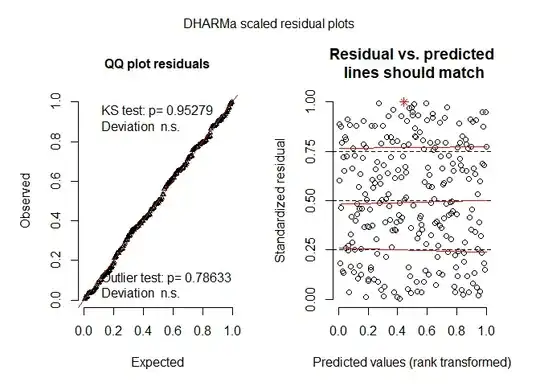
Edit 2 Based on comment suggestions- Patterns look good
ypred = predict(gama_model)
res = residuals(gama_model, type = 'pearson')
plot(ypred,res)
hist(res)
plot(res)
plot(y = res, x = Model_data$Obs)
3. Pearson histograms- based on answer code
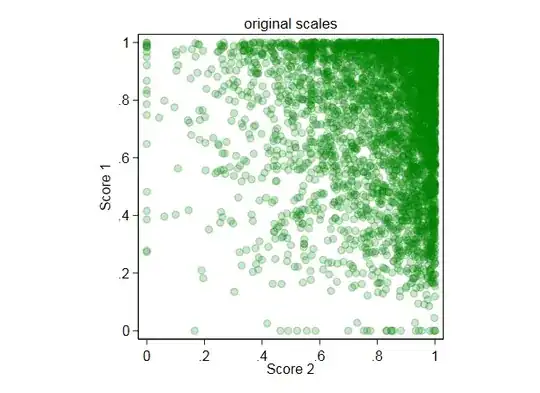
4. Pearson vs pred (link scale)- based on answer code