I am working with a dataset where the response variable looks like an in-between of normal and gamma distribution
Edit: Including model formula and output, as requested, below

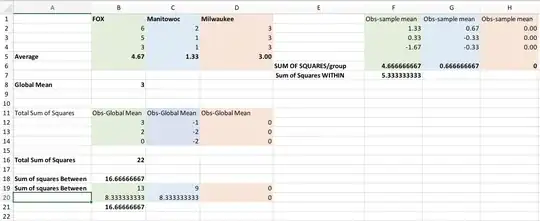
I’m using lme4 because I also have a random effect structure (treatment|farm_id) In calculating AICc values of the null models for both distributions I got - gamma AICc (-158.3) and normal AICc (-61.5)
Implying the gamma is a better fit, correct?
During model verification I’ve run into some issues though. I’ve mainly been following the advice from this post: What are the assumptions of a Gamma GLM or GLMM for hypothesis testing?
GAMMA MODEL VERIFICATION
res.gammaModel <- resid(gammaModel)
qqnorm(res.gammaModel)
qqline(res.gammaModel)›
ypred = predict(gammaModel)
res = residuals(gammaModel, type = 'pearson')
plot(ypred,res)
I feel like the QQplot isn’t great but isn’t bad and the residuals versus predicted aren’t showing patterns
plot(y = res, x = scaled.originalData$treatment)
plot(y = res, x = scaled.originalData$explanatoryVariable)
But then…
plot(y = res.gammaModel, x = originalData$responseVariable)
Here I can’t use x = gammaModel$Obs because its a glmm I guess? And I have some NA values in my data that I needed to get rid of so maybe thats where I went wrong?
Furthermore using DHARMA for the gamma distribution
I’m not really sure where I’m going wrong? I’m using model.sel() and these plots are from the top model (delta AICc only 1 better than the null model).
On the otherhand, the top model of the model.sel() for the normal distribution set of models was the null model (by delta AICc 4) and the DHARMA QQ plot looked better
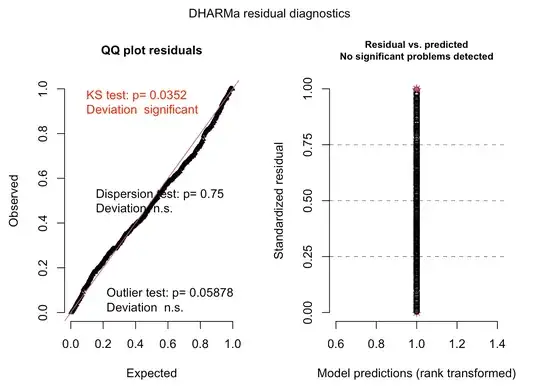 
Any advice would be greatly appreciated!

Any advice would be greatly appreciated!
Edit- Formulas and output - on another note, I realized I am using REML for lmer and ML for glmer - I include the output for an lmer run on ML at the end...
gammalmm <- glmer(response ~ exVar + (treatment|farm_id), data=scaled.originalData, family = Gamma, control=glmerControl(optimizer="bobyqa", optCtrl=list(maxfun=100000)))
Generalized linear mixed model fit by maximum likelihood (Laplace
Approximation) [glmerMod]
Family: Gamma ( inverse )
Formula: response ~ exVar + (treatment | farm_id)
Data: scaled.originalData
AIC BIC logLik deviance df.resid
-160.2394 -120.6222 89.1197 -178.2394 594
Random effects:
Groups Name Std.Dev. Corr
farm_id (Intercept) 0.3859
treatmentBG 0.2714 -0.91
treatmentCC 0.3824 0.12 -0.17
Residual 0.3192
Number of obs: 603, groups: farm_id, 9
Fixed Effects:
(Intercept) exVar
1.20526 0.01206
lmm <- lmer(response ~ 1 + (treatment|farm_id), data=scaled.originalData)
Linear mixed model fit by REML ['lmerMod']
Formula: response ~ 1 + (treatment | farm_id)
Data: scaled.originalData
REML criterion at convergence: -77.7584
Random effects:
Groups Name Std.Dev. Corr
farm_id (Intercept) 0.25875
treatmentBG 0.09519 -0.47
treatmentCC 0.18723 0.00 -0.31
Residual 0.21336
Number of obs: 603, groups: farm_id, 9
Fixed Effects:
(Intercept)
0.7496
Linear mixed model fit by maximum likelihood ['lmerMod']
Formula: response ~ 1 + (treatment | farm_id)
Data: scaled.originalData
AIC BIC logLik deviance df.resid
-65.0964 -29.8811 40.5482 -81.0964 595
Random effects:
Groups Name Std.Dev. Corr
vfarm_id (Intercept) 0.24687
treatmentBG 0.09501 -0.49
treatmentCC 0.18716 0.00 -0.31
Residual 0.21337
Number of obs: 603, groups: farm_id, 9
Fixed Effects:
(Intercept)
0.7495
And the DHARMA for the lmm fit with Maximum likelihood