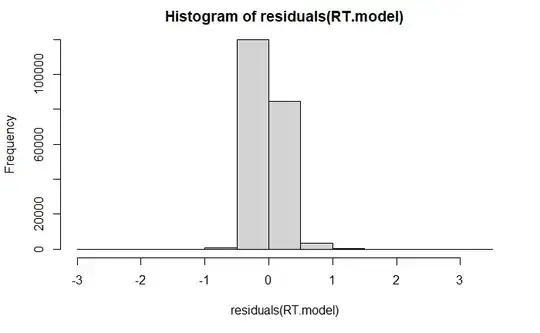
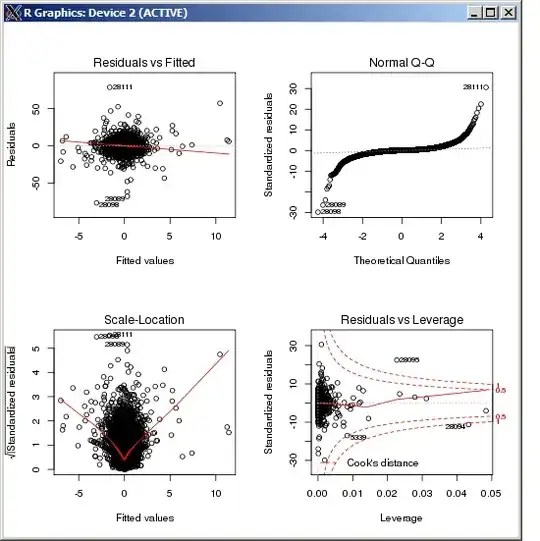
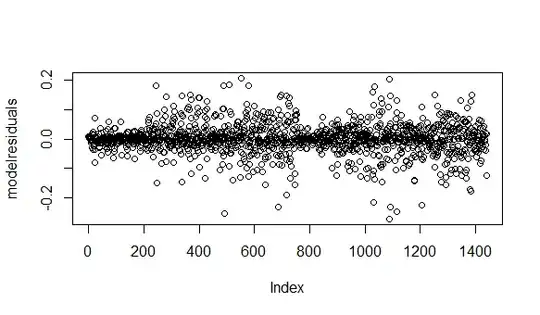
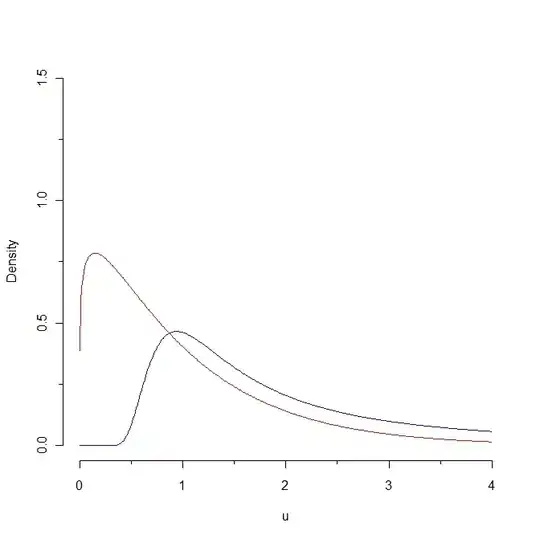
I am running a glmer model
glmer(RT ~ Prob * Bl * Session * Gr + (1 | Participant),
data= Data.trimmed, family = Gamma(link = "log"),
control=glmerControl(optimizer="bobyqa",
optCtrl=list(maxfun=1000000)), nAGQ = 0)
with gamma family and I am trying to determine whether this model shows a good fit. It doesn't seem like it does, but I would like to get someone else's opinion since these models are a lot more complicated to deal with. If you think this is poorly fit, do you have any suggestion for how to improve the model?
I have 209062 rows of data and this is response time data.
I want to determine whether there differences between groups (Gr - 2 levels) on the learning of a task (Pr - 2 levels) across time (within sessions - Bl - 4 levels / across sessions - Session - 2 levels). It doesn't have zero response times, but some close to zero.
I have also added the lmer model at the end.
Glmer Model summary:
Generalized linear mixed model fit by maximum likelihood (Adaptive Gauss-Hermite Quadrature, nAGQ = 0) ['glmerMod']
Family: Gamma ( log )
Formula: RT ~ Probability * Block * Session * Group + (1 | Participant)
Data: Data.trimmed
Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 1e+06))
AIC BIC logLik deviance df.resid
2456107 2456538 -1228012 2456023 209020
Scaled residuals:
Min 1Q Median 3Q Max
-4.297 -0.625 -0.158 0.440 35.691
Random effects:
Groups Name Variance Std.Dev.
Participant (Intercept) 0.002203 0.04694
Residual 0.053481 0.23126
Number of obs: 209062, groups: Participant, 130
Fixed effects:
Estimate Std. Error t value Pr(>|z|)
(Intercept) 6.024e+00 4.182e-03 1440.439 < 2e-16 ***
Probability1 -2.835e-02 7.041e-04 -40.265 < 2e-16 ***
Block2-1 -2.925e-02 2.077e-03 -14.084 < 2e-16 ***
Block3-2 -3.676e-03 2.131e-03 -1.725 0.084500 .
Block4-3 4.085e-03 2.307e-03 1.771 0.076577 .
Block5-4 -1.220e-02 2.380e-03 -5.125 2.98e-07 ***
Session1 4.795e-02 7.323e-04 65.478 < 2e-16 ***
Group1 -4.616e-02 4.182e-03 -11.037 < 2e-16 ***
Probability1:Block2-1 -7.228e-03 2.077e-03 -3.480 0.000501 ***
Probability1:Block3-2 -5.332e-03 2.131e-03 -2.503 0.012331 *
Probability1:Block4-3 -2.076e-02 2.307e-03 -8.999 < 2e-16 ***
Probability1:Block5-4 6.044e-03 2.380e-03 2.539 0.011104 *
Probability1:Session1 1.656e-03 7.046e-04 2.351 0.018743 *
Block2-1:Session1 -1.972e-02 2.077e-03 -9.494 < 2e-16 ***
Block3-2:Session1 -8.521e-03 2.131e-03 -3.999 6.35e-05 ***
Block4-3:Session1 4.380e-05 2.308e-03 0.019 0.984856
Block5-4:Session1 -3.768e-03 2.380e-03 -1.583 0.113389
Probability1:Group1 1.515e-03 7.041e-04 2.151 0.031478 *
Block2-1:Group1 -6.161e-03 2.077e-03 -2.966 0.003015 **
Block3-2:Group1 -1.129e-02 2.131e-03 -5.301 1.15e-07 ***
Block4-3:Group1 7.095e-03 2.307e-03 3.076 0.002101 **
Block5-4:Group1 -4.055e-03 2.380e-03 -1.704 0.088414 .
Session1:Group1 3.782e-03 7.323e-04 5.164 2.41e-07 ***
Probability1:Block2-1:Session1 5.729e-05 2.077e-03 0.028 0.977997
Probability1:Block3-2:Session1 3.543e-03 2.131e-03 1.663 0.096363 .
Probability1:Block4-3:Session1 -6.877e-03 2.308e-03 -2.980 0.002886 **
Probability1:Block5-4:Session1 4.329e-03 2.380e-03 1.819 0.068952 .
Probability1:Block2-1:Group1 -1.238e-03 2.077e-03 -0.596 0.550980
Probability1:Block3-2:Group1 1.022e-02 2.131e-03 4.795 1.63e-06 ***
Probability1:Block4-3:Group1 -6.532e-03 2.307e-03 -2.831 0.004634 **
Probability1:Block5-4:Group1 2.351e-03 2.380e-03 0.988 0.323373
Probability1:Session1:Group1 -1.805e-03 7.046e-04 -2.562 0.010412 *
Block2-1:Session1:Group1 -2.060e-04 2.077e-03 -0.099 0.920984
Block3-2:Session1:Group1 -4.211e-03 2.131e-03 -1.977 0.048094 *
Block4-3:Session1:Group1 3.339e-03 2.308e-03 1.447 0.147888
Block5-4:Session1:Group1 -3.956e-03 2.380e-03 -1.662 0.096539 .
Probability1:Block2-1:Session1:Group1 -1.270e-03 2.077e-03 -0.611 0.540933
Probability1:Block3-2:Session1:Group1 1.678e-03 2.131e-03 0.788 0.430929
Probability1:Block4-3:Session1:Group1 -4.640e-03 2.308e-03 -2.010 0.044392 *
Probability1:Block5-4:Session1:Group1 4.714e-03 2.380e-03 1.980 0.047649 *
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Correlation matrix not shown by default, as p = 40 > 12.
Use print(x, correlation=TRUE) or
vcov(x) if you need it
plotting residuals:
plot(fitted(RT.model), residuals(RT.model))
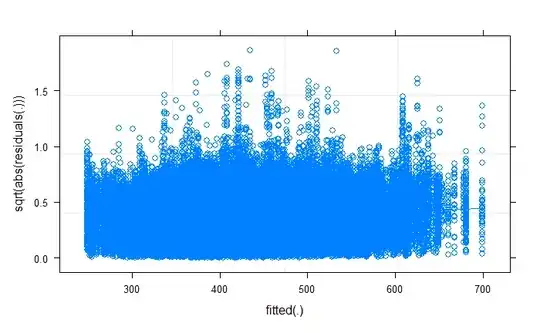
Dharma plot:
plot(RT.model,sqrt(abs(residuals(.))) ~ fitted(.),
type=c("p","smooth"))
For the lmer model:
RT.model <- lmer(logRT ~ Probability * Block * Session * Group + (1 | Participant), data= Data.trimmed)
Linear mixed model fit by REML. t-tests use Satterthwaite's method ['lmerModLmerTest']
Formula: logRT ~ Probability * Block * Session * Group + (1 | Participant)
Data: Data.trimmed
REML criterion at convergence: -51844.2
Scaled residuals:
Min 1Q Median 3Q Max
-23.0325 -0.6283 -0.0801 0.5553 11.0990
Random effects:
Groups Name Variance Std.Dev.
Participant (Intercept) 0.02413 0.1553
Residual 0.04541 0.2131
Number of obs: 209062, groups: Participant, 130
Fixed effects:
Estimate Std. Error df t value Pr(>|t|)
(Intercept) 5.999e+00 1.364e-02 1.283e+02 439.726 < 2e-16 ***
Probability1 -2.731e-02 6.488e-04 2.089e+05 -42.087 < 2e-16 ***
Block2-1 -2.597e-02 1.914e-03 2.089e+05 -13.569 < 2e-16 ***
Block3-2 -6.639e-03 1.963e-03 2.089e+05 -3.381 0.000721 ***
Block4-3 3.532e-03 2.126e-03 2.089e+05 1.662 0.096610 .
Block5-4 -1.598e-02 2.193e-03 2.089e+05 -7.287 3.18e-13 ***
Session1 4.636e-02 6.754e-04 2.090e+05 68.638 < 2e-16 ***
Group1 -4.793e-02 1.364e-02 1.283e+02 -3.513 0.000613 ***
Probability1:Block2-1 -7.919e-03 1.914e-03 2.089e+05 -4.138 3.51e-05 ***
Probability1:Block3-2 -3.684e-03 1.963e-03 2.089e+05 -1.877 0.060567 .
Probability1:Block4-3 -2.211e-02 2.126e-03 2.089e+05 -10.399 < 2e-16 ***
Probability1:Block5-4 6.860e-03 2.193e-03 2.089e+05 3.128 0.001763 **
Probability1:Session1 2.198e-03 6.493e-04 2.089e+05 3.385 0.000711 ***
Block2-1:Session1 -1.879e-02 1.914e-03 2.089e+05 -9.817 < 2e-16 ***
Block3-2:Session1 -7.896e-03 1.963e-03 2.089e+05 -4.022 5.78e-05 ***
Block4-3:Session1 -1.407e-03 2.126e-03 2.089e+05 -0.661 0.508296
Block5-4:Session1 -5.281e-03 2.193e-03 2.089e+05 -2.408 0.016046 *
Probability1:Group1 1.695e-03 6.488e-04 2.089e+05 2.612 0.009010 **
Block2-1:Group1 -7.155e-03 1.914e-03 2.089e+05 -3.739 0.000185 ***
Block3-2:Group1 -1.071e-02 1.963e-03 2.089e+05 -5.453 4.96e-08 ***
Block4-3:Group1 6.027e-03 2.126e-03 2.089e+05 2.835 0.004578 **
Block5-4:Group1 -1.975e-03 2.193e-03 2.089e+05 -0.900 0.367938
Session1:Group1 3.271e-03 6.754e-04 2.090e+05 4.843 1.28e-06 ***
Probability1:Block2-1:Session1 3.932e-05 1.914e-03 2.089e+05 0.021 0.983609
Probability1:Block3-2:Session1 2.946e-03 1.963e-03 2.089e+05 1.500 0.133491
Probability1:Block4-3:Session1 -5.279e-03 2.127e-03 2.089e+05 -2.482 0.013068 *
Probability1:Block5-4:Session1 4.600e-03 2.193e-03 2.089e+05 2.098 0.035950 *
Probability1:Block2-1:Group1 -1.013e-03 1.914e-03 2.089e+05 -0.529 0.596705
Probability1:Block3-2:Group1 1.014e-02 1.963e-03 2.089e+05 5.165 2.41e-07 ***
Probability1:Block4-3:Group1 -6.676e-03 2.126e-03 2.089e+05 -3.140 0.001687 **
Probability1:Block5-4:Group1 2.475e-03 2.193e-03 2.089e+05 1.129 0.259079
Probability1:Session1:Group1 -1.077e-03 6.493e-04 2.089e+05 -1.658 0.097259 .
Block2-1:Session1:Group1 2.446e-03 1.914e-03 2.089e+05 1.278 0.201365
Block3-2:Session1:Group1 -3.793e-03 1.963e-03 2.089e+05 -1.932 0.053342 .
Block4-3:Session1:Group1 1.633e-03 2.126e-03 2.089e+05 0.768 0.442515
Block5-4:Session1:Group1 -2.495e-03 2.193e-03 2.089e+05 -1.137 0.255393
Probability1:Block2-1:Session1:Group1 -2.243e-03 1.914e-03 2.089e+05 -1.172 0.241206
Probability1:Block3-2:Session1:Group1 1.190e-03 1.963e-03 2.089e+05 0.606 0.544432
Probability1:Block4-3:Session1:Group1 -3.378e-03 2.127e-03 2.089e+05 -1.588 0.112179
Probability1:Block5-4:Session1:Group1 5.139e-03 2.193e-03 2.089e+05 2.343 0.019126 *
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Correlation matrix not shown by default, as p = 40 > 12.
Use print(x, correlation=TRUE) or
vcov(x) if you need it
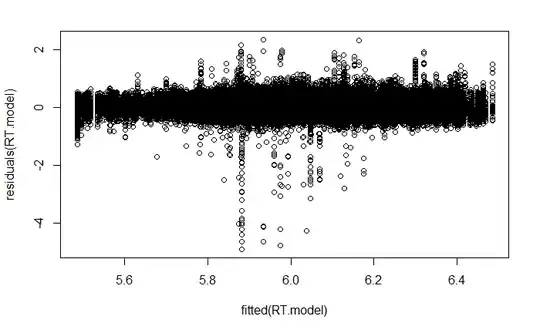
plotting residuals
plot(fitted(RT.model),residuals(RT.model))
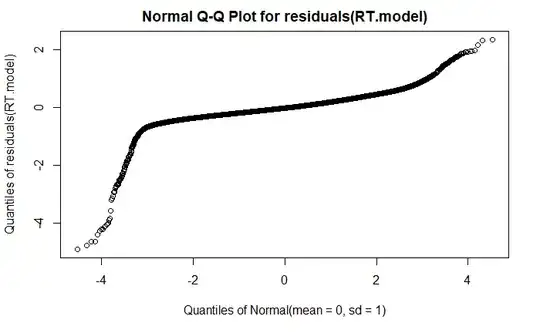
qqplot
DHARMa plot