I have data that looks like the following
>df
id response Factor Count1 Count2
1 0 A 6 1
1 0 B 5 2
1 0 C 4 3
2 1 A 3 4
2 1 C 2 5
2 1 D 1 6
The goal is to see how the Factor and the Count covariates effect the response.
My current set up is glm(response~Factor+Count1+Count2,family=binomial(link=logit))
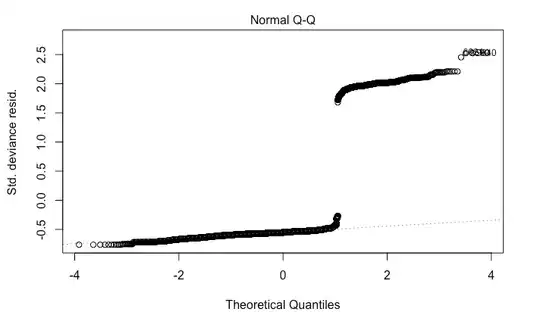
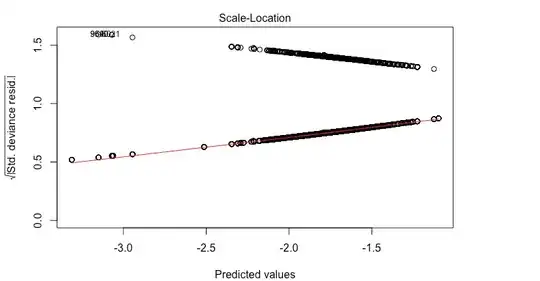
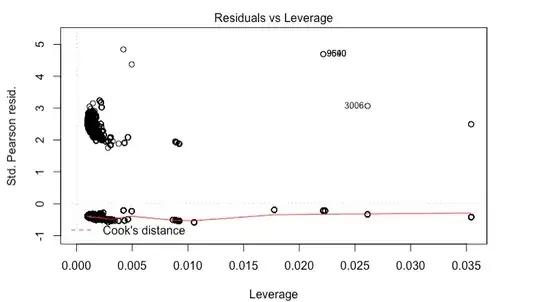
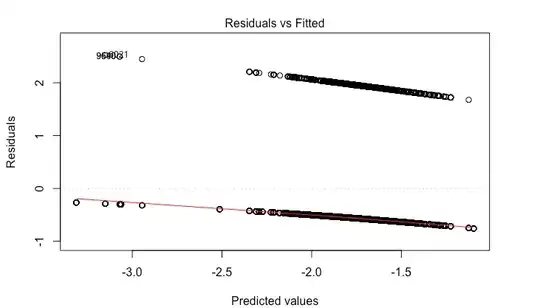
The problem is that I am getting wild residual plots.
I am wondering if i should try running a mixed effects model since I have multiple observations for each id. I also wonder if my ratio of 1s to 0s in the response are to great. I have 11,000 rows and 1,500 are 1s.
Any insight would be helpful. Thanks!
Edit: I just came across this post about each id being drawn from a Bernoulli. It turns out that I was able to find a natural grouping for my ids and make proper proportions from there. My Normal Q-Q plot is not great, but at least they don't have a large gap in them anymore.