NB the deviance (or Pearson) residuals are not expected to have a normal distribution except for a Gaussian model. For the logistic regression case, as @Stat says, deviance residuals for the $i$th observation $y_i$ are given by
$$r^{\mathrm{D}}_i=-\sqrt{2\left|\log{(1-\hat{\pi}_i)}\right|}$$
if $y_i=0$ &
$$r^{\mathrm{D}}_i=\sqrt{2\left|\log{(\hat{\pi}_i)}\right|}$$
if $y_i=1$, where $\hat{\pi_i}$ is the fitted Bernoulli probability. As each can take only one of two values, it's clear their distribution cannot be normal, even for a correctly specified model:
#generate Bernoulli probabilities from true model
x <-rnorm(100)
p<-exp(x)/(1+exp(x))
#one replication per predictor value
n <- rep(1,100)
#simulate response
y <- rbinom(100,n,p)
#fit model
glm(cbind(y,n-y)~x,family="binomial") -> mod
#make quantile-quantile plot of residuals
qqnorm(residuals(mod, type="deviance"))
abline(a=0,b=1)
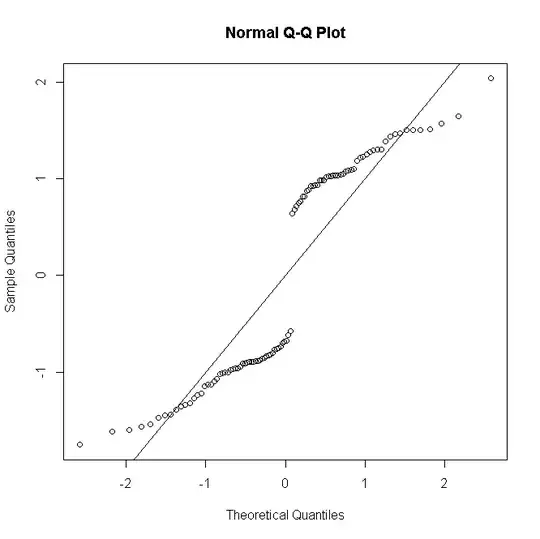
But if there are $n_i$ replicate observations for the $i$th predictor pattern, & the deviance residual is defined so as to gather these up
$$r^{\mathrm{D}}_i=\operatorname{sgn}({y_i-n_i\hat{\pi}_i})\sqrt{2\left[y_i\log{\frac{y_i}{n\hat{\pi}_i}} + (n_i-y_i)\log{\frac{n_i-y_i}{n_i(1-\hat{\pi}_i)}}\right]}$$
(where $y_i$ is now the count of successes from 0 to $n_i$) then as $n_i$ gets larger the distribution of the residuals approximates more to normality:
#many replications per predictor value
n <- rep(30,100)
#simulate response
y<-rbinom(100,n,p)
#fit model
glm(cbind(y,n-y)~x,family="binomial")->mod
#make quantile-quantile plot of residuals
qqnorm(residuals(mod, type="deviance"))
abline(a=0,b=1)
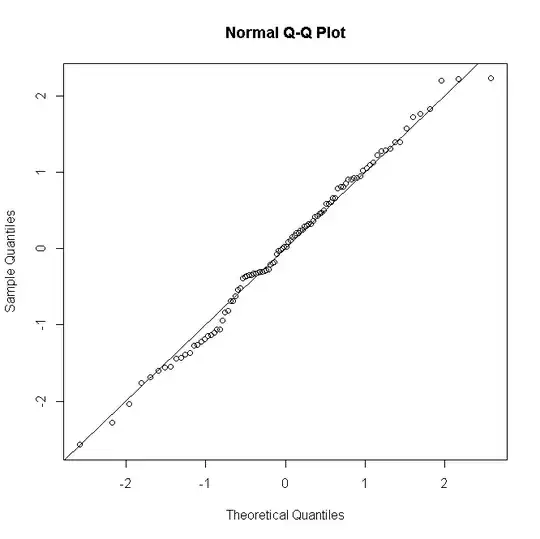
Things are similar for Poisson or negative binomial GLMs: for low predicted counts the distribution of residuals is discrete & skewed, but tends to normality for larger counts under a correctly specified model.
It's not usual, at least not in my neck of the woods, to conduct a formal test of residual normality; if normality testing is essentially useless when your model assumes exact normality, then a fortiori it's useless when it doesn't. Nevertheless, for unsaturated models, graphical residual diagnostics are useful for assessing the presence & the nature of lack of fit, taking normality with a pinch or a fistful of salt depending on the number of replicates per predictor pattern.

