I am looking to group/merge nodes in a graph using graph clustering in 'r'.
Here is a stunningly toy variation of my problem.
- There are two "clusters"
- There is a "bridge" connecting the clusters
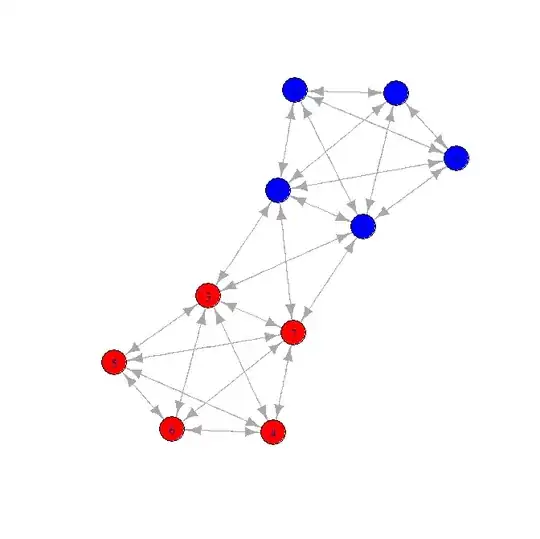
Here is a candidate network:
When I look at the connection distance, the "hopcount", if you will, then I can get the following matrix :
mymatrix <- rbind(
c(1,1,2,3,3,3,2,1,1,1),
c(1,1,1,2,2,2,1,1,1,1),
c(2,1,1,1,1,1,1,1,2,2),
c(3,2,1,1,1,1,1,2,3,3),
c(3,2,1,1,1,1,1,2,3,3),
c(3,2,1,1,1,1,1,2,2,2),
c(2,1,1,1,1,1,1,1,2,2),
c(1,1,1,2,2,2,1,1,1,1),
c(1,1,2,3,3,2,2,1,1,1),
c(1,1,2,3,3,2,2,1,1,1))
Thoughts here:
- By luck or due to the simplicity of the toy the matrix has obvious patches this is not going to be the case in the (very large) matrix. If I randomized the relationship between point and row then it would not be so clean.
- I might have got one wrong - so if I have a typo, let me know.
- Hop-count here is shortest number of hops to connect point on row i with point on column j. A self-hop is still a hop, so the diagonal is all ones.
So in this matrix larger distance (hops) has a higher number. If I wanted a matrix showing "connectivity" instead of distance, then I could do a dot-inverse, where each cell of the matrix is replaced with its multiplicative inverse.
Questions:
To help me find my own way:
- What are the terms for reducing the number of nodes on a graph by combining them? Is it clustering, merging, munging - what are the words that I should use?
- What are the proven techniques? Is there a textbook on the topic? Can you point to papers or websites?
- Now I tried to look here first - it is a great "first check" spot. I didn't find what I was looking for. If I missed it (not unlikely) can you point me to an answered question or two on the topic here at CV?
To get me where I am going:
- Is there an 'R' package that will properly cluster the nodes on the network?
- Could you point me to example code to do this?
- Is there an 'R' package that will graphically present the resulting reduced network?
- Could you point me to example code to do this?
Thanks in advance.