I am practicing Mixture of Gaussians and found the below dataset snoq, which is the precipitation amounts recorded at a US region, with NA and no precipitation days removed.
snoqualmie <- read.csv("http://www.stat.cmu.edu/~cshalizi/402/lectures/16-glm-practicals/snoqualmie.csv",header=FALSE)
snoqualmie.vector <- na.omit(unlist(snoqualmie)) # remove NA's and flatten
snoq <- snoqualmie.vector[snoqualmie.vector > 0] # days where precipitation was greater than 0
In the exercise code, the instructor fits a mixture of 2 Gaussians to the data with the below code (the plot.normal.components function is given at the end of my question):
if(!require("mixtools")) { install.packages("mixtools"); require("mixtools") }
snoq.k2 <- normalmixEM(snoq, k=2, maxit=100, epsilon=0.01)
plot(hist(snoq,breaks=101),col="grey",border="grey",freq=FALSE,
xlab="Precipitation (1/100 inch)",main="Precipitation in Snoqualmie Falls")
lines(density(snoq),lty=2)
sapply(1:2,plot.normal.components,mixture=snoq.k2)
The dotted line is the kernel density estimation (on the empirical pdf) and the two black curves are the fitted Gaussians.
Now, I thought the distribution of snoq is similar to that of an exponential distribution, so my first instict here was to log transform the data and then investigate what happens if I try to fit a Mixture of Gaussians on log data rather than the raw data as the instructor did:
log_snoq <- log(snoq)
log_snoq.k3 = normalmixEM(log_snoq, k=3, maxit=100, epsilon=0.01) # does not converge!
plot(hist(log_snoq,breaks=101),col="grey",border="grey",freq=FALSE,
xlab="Log Precipitation (1/100 inch)",main="Precipitation in Snoqualmie Falls")
lines(density(log_snoq),lty=2)
sapply(1:3,plot.normal.components,mixture=log_snoq.k3)
I chose k=3 in this case, just because the kernel density estimator showed 3 bumps on the dotted line (in the figure below), which I interpreted as three local maxima that can be modelled by 3 univariate Gaussians. However, during EM I get a warning that says WARNING! NOT CONVERGENT!, and the resulting Gaussian components look as below: (I do realise that the log transform does not produce a beautifully Gaussian-looking distribution, but the raw data itself looks less suitable to be modelled with a (mixture of) Gaussian(s) to me anyway.)
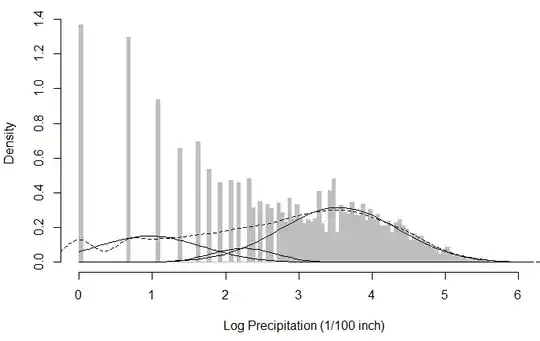
My question is, in this particular example is it wrong to log-transform? Does this in general complicate the application of Mixture of Gaussians? Any recommendations / comments are welcome!
The function plot.normal.components:
plot.normal.components <- function(mixture,component.number,...) {
curve(mixture$lambda[component.number] *
dnorm(x,mean=mixture$mu[component.number],
sd=mixture$sigma[component.number]), add=TRUE, ...)
}