Suppose I have normally distributed data. For each element of the data I want to check how many SDs it is away from the mean. There might be an outlier in the data (likely only one, but might be also two or three) or not, but this outlier is basically what I am looking for. Does it make sense to temporarily exclude the element I am currently looking at from the calculation of the mean and the SD? My thinking is that if it is close to the mean, it does not have any impact. If it is an outlier, it might bias the calculation of mean and SD and lower the probability that it is detected. I am not a statistician, so any help is appreciated!
-
8It makes perfect sense and is the basis for many outlier-detection techniques. But rather than inventing your own method, which might or might not work (and the latter is far more likely even with methods newly invented by statisticians, which is why they need careful study), why don't you use one that has been theoretically checked and empirically tested? – whuber Oct 22 '14 at 15:42
-
Thanks for pointing that out. I will look up those techniques and see whether they perform well on my data! – Oliver Oct 22 '14 at 16:07
-
1Check out this page on Regression Deletion Diagnostics in R: http://stat.ethz.ch/R-manual/R-patched/library/stats/html/influence.measures.html – Ben Ogorek Oct 23 '14 at 05:10
-
....And [this](http://stats.stackexchange.com/a/50780/603) answer for illustration of why they can t be depended upon to find more than a single outlier. – user603 Oct 23 '14 at 09:08
-
Great thoughts above on the idea of flagging outliers. Sometime back, I had written an article on the idea of loss-pass filters on flagging anomalies. Hope this helps in extending the idea presented above. Link to the article: https://www.datascience.com/blog/python-anomaly-detection – Pramit Feb 21 '19 at 18:42
1 Answers
It might seem counter-intuitive, but using the approach you describe doesn't make sense (to take your wording, I would rather write "can lead to outcomes very different from those intended") and one should never do it: the risks of it not working are consequential and besides, there exists a simpler, much safer and better established alternative available at no extra cost.
First, it is true that if there is a single outlier, then you will eventually find it using the procedure you suggest. But, in general (when there may be more than a single outlier in the data), the algorithm you suggest completely breaks down, in the sense of potentially leading you to reject a good data point as an outlier or keep outliers as good data points with potentially catastrophic consequences.
Below, I give a simple numerical example where the rule you propose breaks down and then I propose a much safer and more established alternative, but before this I will explain a) what is wrong with the method you propose and b) what the usually preferred alternative to it is.
In essence, you cannot use the distance of an observation from the leave one out mean and standard deviation of your data to reliably detect outliers because the estimates you use (leave one out mean and standard deviation) are still liable to being pulled towards the remaining outliers: this is called the masking effect.
In a nutshell, one simple way to reliably detect outliers is to use the general idea you suggested (distance from estimate of location and scale) but replacing the estimators you used (leave one out mean, sd) by robust ones--i.e., estimates designed to be much less susceptible to being swayed by outliers.
Consider this example, where I add 3 outliers to 47 genuine observations drawn from a Normal 0,1:
n <- 50
set.seed(123) # for reproducibility
x <- round(rnorm(n,0,1), 1)
x[1] <- x[1]+1000
x[2] <- x[2]+10
x[3] <- x[3]+10
The code below computes the outlyingness index based on the leave one out mean and standard deviation (e.g. the approach you suggest).
out_1 <- rep(NA,n)
for(i in 1:n){ out_1[i] <- abs( x[i]-mean(x[-i]) )/sd(x[-i]) }
and this code produces the plot you see below.
plot(x, out_1, ylim=c(0,1), xlim=c(-3,20))
points(x[1:3], out_1[1:3], col="red", pch=16)
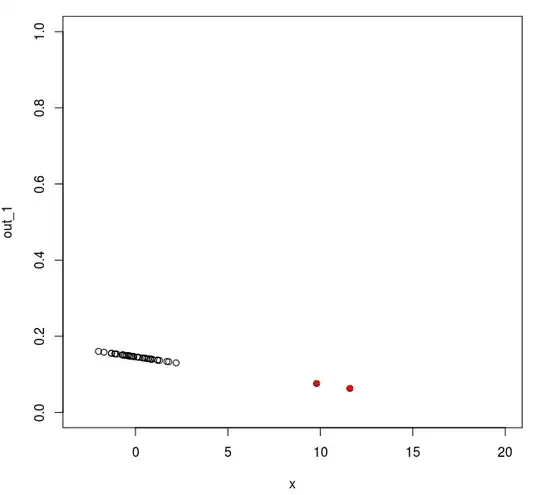
Image 1 depicts the value of your outlyingness index as a function of the value of the observations (the furthest away of the outliers is outside the range of this plot but the other two are shown as red dots). As you can see, except for the most extreme one, an outlyingness index constructed as you suggest would fail to reveal the outliers: indeed the second and third (milder) outliers now even have a value (on your outlyingness index) smaller than all the genuine observations!...Under the approach you suggest, one would keep these two extreme outliers in the set of genuine observations, leading you to use the 49 remaining observations as if they were coming from the same homogeneous process, giving you a final estimate of the mean and sd based on these 49 data points of 0.45 and 2.32, a very poor description of either part of your sample!
Contrast this outcome with the results you would have obtained using an outlier detection rule based on the median and the mad where the outlyingness of point $x_i$ wrt to a data vector $X$ is
$$O(x_i,X)=\frac{|x_i-\mbox{med}(X)|}{\mbox{mad}(X)}$$
where $\mbox{med}(X)$ is the median of the entries of $X$ (all of them, without exclusion) and $\mbox{mad}(X)$ is their median absolute deviation times 1.4826 (I defer to the linked wiki article for an explanation of where this number comes from since it is orthogonal to the main issue here).
In R, this second outlyingness index can be computed as:
out_2 <- abs( x-median(x) )/mad(x)
and plotted (as before) using:
plot(x, out_2, ylim=c(0,15), xlim=c(-3,20))
points(x[1:3], out_2[1:3], col="red", pch=16)

Image 2 plots the value of this alternative outlyingness index for the same data set. As you can see, now all three outliers are clearly revealed as such. Furthermore, this outlier detection rule has some established statistical properties. This leads, among other things, to usable cut-off rules. For example, if the genuine part of the data can be assumed to be drawn from a symmetric distribution with finite second moment, you can reject all data points for which
$$\frac{|x_i-\mbox{med}(X)|}{\mbox{mad}(X)}>3.5$$
as outliers. In the example above, application of this rule would lead you to correctly flag observation 1,2 and 3. Rejecting these, the mean and sd of the remaining observations is 0.021 and 0.93 receptively, a much better description of the genuine part of the sample!
- 21,225
- 3
- 71
- 135
-
2+1 despite the first sentence, which you immediately contradict (the OP's proposal *does* make sense when at most one outlier is assumed; your objection concerns problems with this procedure when that assumption is violated). – whuber Oct 22 '14 at 17:22
-
@whuber : Indeed "make sense" is so broad a characterization as to be useless as an assessment for any methodology. But given the risks (the OP maintains that there could be more than one outliers) the naive solution doesn't make sense from a risk/pay off point of view: the risk are non 0 and I hardly see a pay off over the more established solution I discuss. – user603 Oct 22 '14 at 17:25
-
1Thank you. In the meantime I deleted my previous comment, anticipating it will become obsolete after your edits. – whuber Oct 22 '14 at 17:33
-
3The phenomenon where several outliers make single-outlier-detection blind to any of them is often called *masking*. This may help people to locate more information relating to the issue. – Glen_b Oct 23 '14 at 00:40
-
1@user603 Nice job creating an illustrative scenario but I think you're throwing out the baby with the bathwater. Regression deletion diagnostics aren't perfect but they are widely applicable and have stood the test of time. Taking the median is fine but I wonder how you'd extend your approach to more complex likelihood based models. – Ben Ogorek Oct 23 '14 at 05:21
-
@BenOgorek: 'stood the test of time': the approach illustrated in my example (using the MAD/median rule) was already used by Gauss around 1813. 'More complicated models': sure: you might want to check a recent textbook on the [subject](http://www.amazon.com/Robust-Statistics-Ricardo-A-Maronna/dp/0470010924/ref=pd_bxgy_b_img_y) – user603 Oct 23 '14 at 07:59
-
I do want to learn more about robust linear models, and thanks for the resource. I'm linking to one myself, "The Changing History of Robustness," by Stephen Stigler: https://files.nyu.edu/ts43/public/research/Stigler.pdf. According to Stigler, the field of Robust Statistics fully emerged in the 60s and seemed like it would be the new paradigm in the 70s, but it just didn't happen. He argues that the properties of, say, Least Squares, are just too appealing to give up. – Ben Ogorek Oct 23 '14 at 14:47
-
@BenOgorek: I think of robust statistics in a dispassionate way, like I would of any other tool. Knowing what I know about the 'appealing properties of LS', I still wouldn t have used it in a situation like that illustrated above, for the reasons I tried to explain, just like I wouldn t use a fork to eat soup, despite the appealing properties of forks. Maybe Stigler would have, but it is immaterial to me. I really enjoyed reading his books tho and recommend them warmly to anyone interested in the history of statistics. – user603 Oct 23 '14 at 14:58
-
2+6, This is a really great answer--clearly & thoroughly explained, illustrated with code, figures & formulas. I tweaked the code formatting slightly to make it a little easier to read. If you don't like it, roll it back w/ my apologies. – gung - Reinstate Monica Oct 24 '14 at 21:50
-
@user603 great answer. If I want to sample a value from such a univariate distribution, would a gaussian sample using median (in place of mean) and MAD (in place of stad deviation) be a better/robust sampler? – Abhinav Dec 03 '15 at 16:02
-
@Abhinav sorry for the late answer. What do you mean by a "Gaussian sample"? – user603 Dec 14 '15 at 17:41
-
@user603 I mean, sampling a value from distribution N(median, MAD) is better than N(mean, std-dev) ? Or this does not make sense? – Abhinav Dec 15 '15 at 16:30