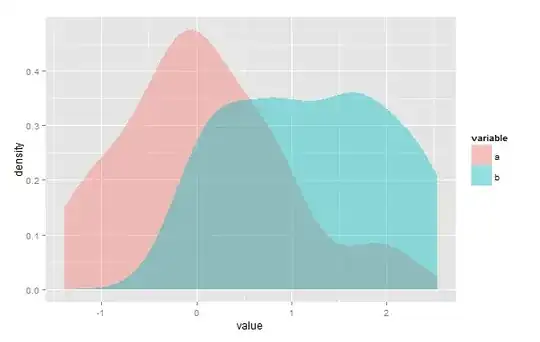
An alternative method for empirical estimation is to use the ROC (Receiver Operating Curve) technology for the estimation. The Youden threshold gives us an empirical estimate for the main point of intersection (see https://journals.lww.com/epidem/Fulltext/2005/01000/Optimal_Cut_point_and_Its_Corresponding_Youden.11.aspx and https://math.stackexchange.com/questions/2404750/intersection-normal-distributions-and-minimal-decision-error/2435957#2435957).
The Youden threshold is the threshold where the sum of test sensitivity and specificity is maximized and the sum of the error rates (False positive Rate and False Negative Rate) is minimized. The overlap is equal to this minimal sum of error rates.
library(UncertainInterval)
simple_roc2 <- function(ref, test){
tab = table(test, ref) # head(tab)
data.frame(threshold=paste('>=',rownames(tab)),
ref0 = tab[,1],
ref1 = tab[,2],
FPR = rev(cumsum(rev(tab[,1])/sum(tab[,1]))), # 1-Sp
TPR = rev(cumsum(rev(tab[,2])/sum(tab[,2]))), # Se
row.names=1:nrow(tab))
}
a <- rnorm(10000)
b <- rnorm(10000, 2)
test=c(a,b)
ref=c(rep(0, length(a)), rep(1, length(b)))
# table(test, ref)
res = simple_roc2(ref, test)
res$FNR = 1-res$TPR # 1-Se
pos.optimal.threshold = which.min(res$FPR+res$FNR)
optimal.threshold=row.names(table(test, ref))[pos.optimal.threshold] # Youden threshold
plotMD(ref, test) # library(UncertainInterval) # includes kernel intersection estimate
abline(v=optimal.threshold, col='red')
overlap1(a, b)
(overlap2 = min(res$FPR+res$FNR))
In this case, this non-parametric estimate has a slight tendency of under estimation of the true value. This roc-technique only handles a single (main) point of intersection. It is not dependent on any specific distribution. Make sure that distribution b has the higher values (mean(b) > mean(a)).
Repeatedly eyeballing plots produced by plotMD shows that with 2 * 100 cases the sample overlap differs considerably. Most differences are due to sample differences, but, dependent on the distributions, all methods have conditions in which they do not work properly. Using a gaussian kernel density is sensitive to spikes in the data, which can be underestimated. Kernel density methods are dependent on the fine-tuning parameters given to the density function. The roc-method has no parameters, but it assumes a single point of intersection. Consequentially, it may overestimate overlap when an additional point of intersection is present (the critical point is the presence of more than one point of intersection, not variance). This overestimation may be negligible when this secondary point of intersection is at the tails of both distributions.
How to make sense of the different methods and suggestions? Devising a test is most simple when we know the true value of two distributions.
The true value of the overlap for the two normal distributions is easy to calculate. The point of intersection is simply the mean of the two means of the distributions, as they have equal variance: 1. The true overlap is then 0.3173105:
(true.overlap = pnorm(1,2,1)+ 1-pnorm(1,0,1))
See https://stackoverflow.com/questions/16982146/point-of-intersection-2-normal-curves/45184024#45184024 for a general method to calculate the point of intersection for two normal distributions.
In the original problem, there is a mix of a normal and a uniform distribution. The true value is in that case:
true.value=sum(pmin(diff(pnorm(0:3)),1/3))
Running a simulation can show us which estimation method produces estimates that are closest to the true value:
library(sfsmisc)
overlap1 <- function(a,b){
lower <- min(c(a, b)) - 1
upper <- max(c(a, b)) + 1
# generate kernel densities
da <- density(a, from=lower, to=upper)
db <- density(b, from=lower, to=upper)
d <- data.frame(x=da$x, a=da$y, b=db$y)
# calculate intersection densities
d$w <- pmin(d$a, d$b)
# integrate areas under curves
total <- integrate.xy(d$x, d$a) + integrate.xy(d$x, d$b)
intersection <- integrate.xy(d$x, d$w)
# compute overlap coefficient
2 * intersection / total
}
library(overlap)
library(scales)
# For explanation of the next function see the answer of S. Venne
overlapEstimates =function(a, b){
a = data.frame( value = a, Source = "a" )
b = data.frame( value = b, Source = "b" )
d = rbind(a, b)
d$value <- scales::rescale( d$value, to = c(0,2*pi) )
a <- d[d$Source == "a", 1]
b <- d[d$Source == "b", 1]
overlapEst(a, b, kmax = 3, adjust=c(0.8, 1, 4), n.grid = 500)
}
nsim=1000; nobs=100; m1=4; sd1=1; m2=6; sd2=1; poi=5
(true.overlap= 1-pnorm(poi, m1, sd1)+pnorm(poi,m2,sd2))
out=matrix(NA,nrow=nsim,ncol=4)
set.seed(0)
for (i in 1:nsim){
x <- rnorm( nobs, m1, sd1 )
y <- rnorm( nobs, m2, sd2 )
out[i,1] = overlap1(x,y)
out[i,2] = overlapping::overlap(list( x = x, y = y ))$OV
out[i,3] = overlapEstimates(x,y)['Dhat4']
out[i,4] = roc.overlap(x,y)
}
(true.overlap=pnorm(poi,m2,sd2)+1-pnorm(poi,m1,sd1))
colMeans(out-true.overlap) # estimation errors
apply(out, 2, sd) # # sd of the estimation errors
apply(out, 2, range)-true.overlap
par(mfrow=c(2,2))
br = seq(-.33,+.33,by=0.05)
hist(out[,1]-true.overlap, breaks=br, ylim=c(0,500),
xlim=c(-.33,.33), main='overlap1');
abline(v=0, col='red')
hist(out[,2]-true.overlap, breaks=br, ylim=c(0,500),
xlim=c(-.33,.33), main='overlapping::overlap')
abline(v=0, col='red')
hist(out[,3]-true.overlap, breaks=br, ylim=c(0,600),
xlim=c(-.33,.33), main='overlap::overlapEst')
abline(v=0, col='red')
hist(out[,4]-true.overlap, breaks=br, ylim=c(0,500),
xlim=c(-.33,.33), main="ROC estimate");
abline(v=0, col='red')
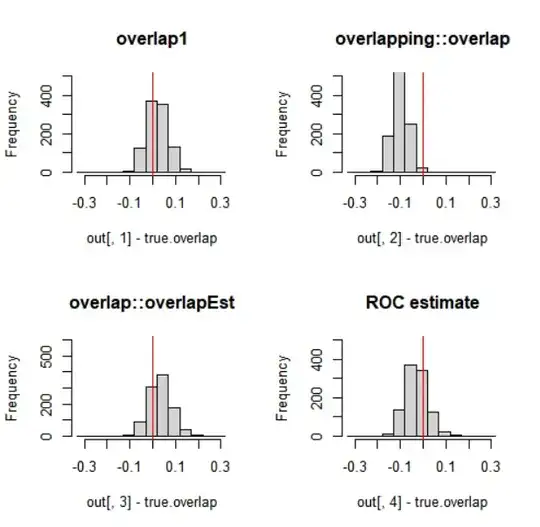
In this case, especially the function overlapping::overlap has a tendency of (slight) under-estimation, while overlap1 shows the least estimation error. Estimates that use the density function in one way or another, can produce better or worse results dependent on the parameters given to the density function. The roc method does not have parameters, which can be an advantage.
It is always wise to carefully look at a plot of the overlapping distributions and to devise a relevant testing method whether the technique for overlap estimation works as expected for the kind of data that you have. Especially techniques that systematically produce estimates that are often too low or to high are better not used.