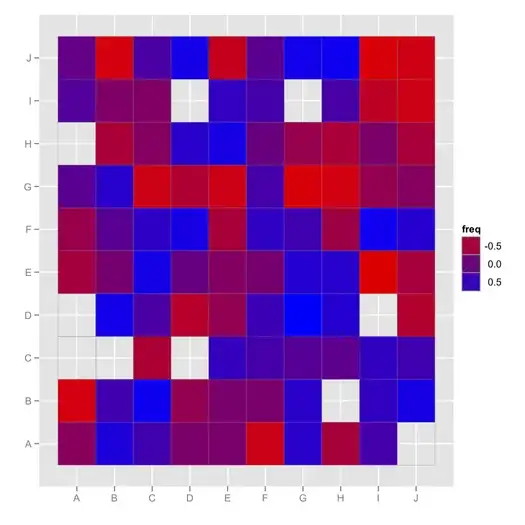
I'd like to obtain a graphic representation of the correlations in articles I have gathered so far to easily explore the relationships between variables. I used to draw a (messy) graph but I have too much data now.
Basically, I have a table with:
- [0]: name of variable 1
- [1]: name of variable 2
- [2]: correlation value
The "overall" matrix is incomplete (e.g., I have the correlation of V1*V2, V2*V3, but not V1*V3).
Is there a way to graphically represent this ?