Can anyone explain why the BP, Breusch-Pagan, test rejects homoscedasticity with such an apparently randomized plot of residuals?
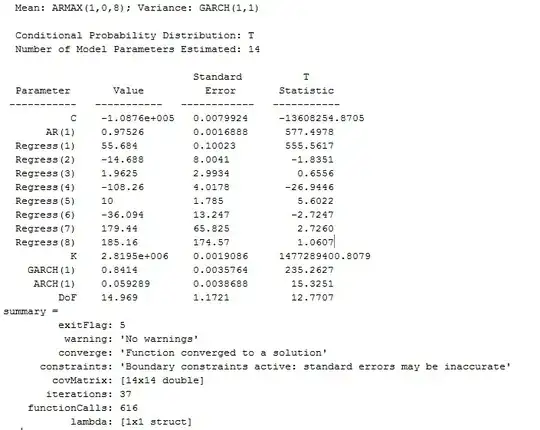

Can anyone explain why the BP, Breusch-Pagan, test rejects homoscedasticity with such an apparently randomized plot of residuals?


You have drawn the wrong scatterplot, I believe.
You should have a scatterplot of the residuals against the fitted values from the model, not "index" which is just the order in which the observations went into the data set