I re-edited this question for clarity:
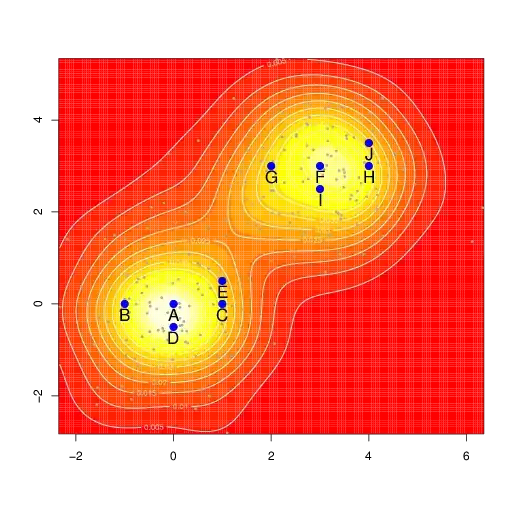
- I have coordinates obtained by tracking birds with GPS (all points in plot).
- I've used these points to perform kernel density estimation to reveal the areas with most dense bird activity (areas "1" and "2" in plot).
- I also know coordinates of nesting trees where each nest was protected by female and male (big blue points with labels ("A" ..."J").
- On each nest were done nest-defense recordings (10 min each recording, on three various intruders, twice per the same intruder)= 60 min total for each nest.
- Each bird (female and male) exhibit 4 behaviours: "attack", "threat", "jump","check". I have all these values (duration).
Thanks to David Robinson for his time. I've done mistake in refer! I want to refer to Table 2. in this article:
onlinelibrary.wiley.com/doi/10.1111/jbi.12048/full
In article is used analysis of molecular variance (AMOVA). May I use something similar to my data in order to explain: "source of variation"
- among areas
- among nesting trees within clusters
- among individuals (female vs. male) within nesting tree
Is it a problem that this AMOVA is (probably) "designed" on genetic data? Which procedure in appropriate for my data? Or should I use nested ANOVA? How?
Thanks.