I am trying to fit my data to the one of the continuous PDF (I suggest it to be gamma- or lognormal-distributed). The data consists of about 6000 positive floats. But the results of the Kolmogorov-Smirnov test completely refute my expectations providing the very low p-values.
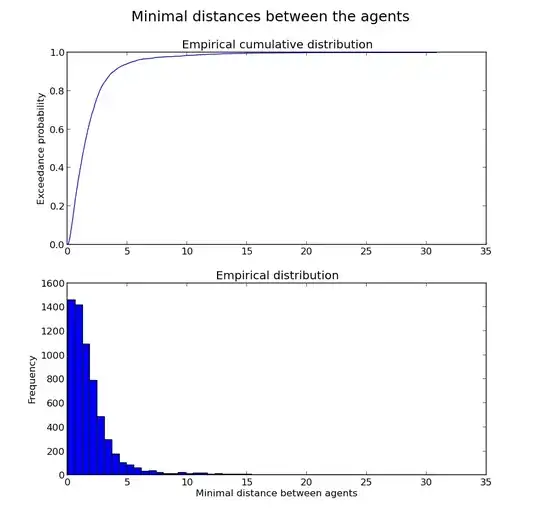
Data empirical distribution
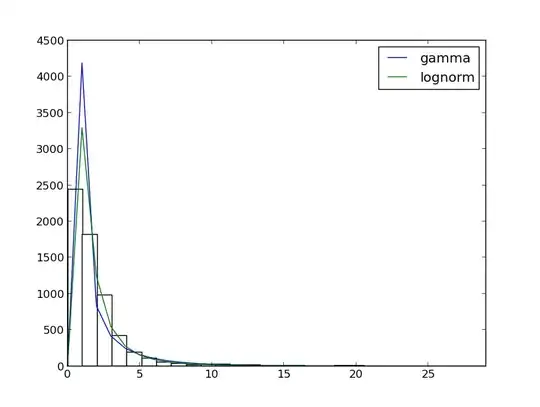
Distribution fitting
Python code:
import numpy
import sys
import json
import matplotlib.pyplot as plt
import scipy
from scipy.stats import *
dist_names = ['gamma', 'lognorm']
limit = 30
def distro():
#input file
with open(sys.argv[1]) as f:
y = numpy.array(json.load(f))
#output
results = {}
size = y.__len__()
x = scipy.arange(size)
h = plt.hist(y, bins=limit, color='w')
for dist_name in dist_names:
dist = getattr(scipy.stats, dist_name)
param = dist.fit(y)
goodness_of_fit = kstest(y, dist_name, param)
results[dist_name] = goodness_of_fit
pdf_fitted = dist.pdf(x, *param) * size
plt.plot(pdf_fitted, label=dist_name)
plt.xlim(0, limit-1)
plt.legend(loc='upper right')
for k, v in results.iteritems():
print(k, v)
plt.show()
This is the output:
- p-value is almost 0
'lognorm', (0.1111486360863001, 1.1233698406822002e-66) - p-value is 0
'gamma', (0.30531260123096859, 0.0)
Does it mean that my data does not fit gamma distribution?.. But they seem so similar...