I have two sets of data of protein-protein interactions in a matrices entitled: s1m and s2m. Each DB and AD pair make an interaction and the one matrix looks like:
> head(s1m)
DB_num AD_num
[1,] 2 8153
[2,] 7 3553
[3,] 8 4812
[4,] 13 7838
[5,] 24 3315
[6,] 24 6012
I can then plot the density of the points basically showing where the points are the most concentrated:
s1m:
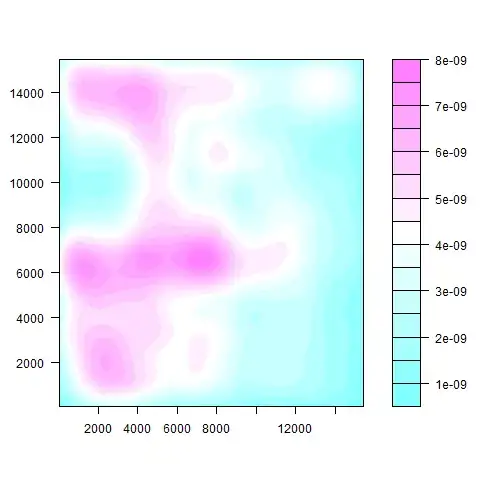
s2m:
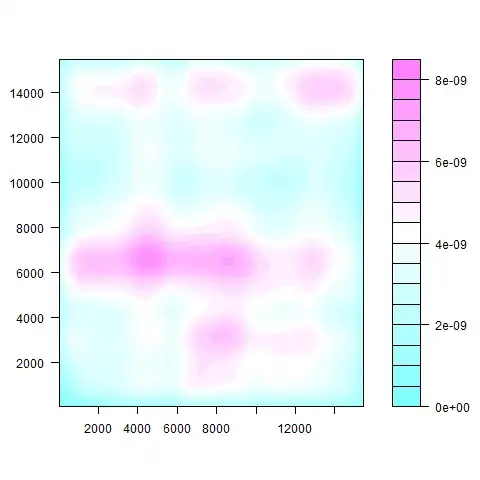
The code I used in R to make these plots was:
z <- kde2d(s1m[,1], s1m[,2], n=50)
filled.contour(z)
z <- kde2d(s2m[,1], s2m[,2], n=50)
filled.contour(z)
I want to be able to somehow compare how simiarly these plots are rather than just looking at them by eye. Is there someway to do this? By the way, I know very little about statistics. These are very large datasets also, something like 10,000 points among a matrix of 15k by 15k.