I am trying to determine parameters in a bayesian network with two latent variables (in blue).
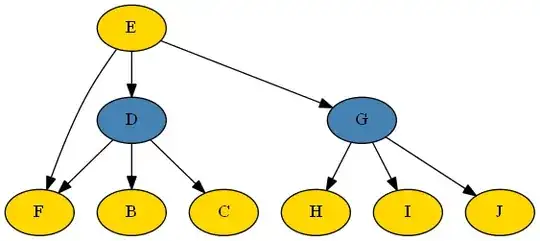
Every variable is discrete with 2-4 categories. The latent variables have 3 categories each.
I am trying to find an R package that will let me define the DAG and then learn the conditional probability parameters from data:
B C D E F G H I J
2 1 ? 3 2 ? 1 4 4
2 2 ? 3 2 ? 1 3 2
2 1 ? 3 2 ? 1 4 2
3 3 ? 3 2 ? 1 4 2
3 1 ? 3 2 ? 1 1 4
3 3 ? 3 2 ? 1 4 1
...
I have tried working with:
bnlearn: as far as I can tell the parameter estimation doesn't support latent variablescatnet: I can't really tell what is going on here, but the results haven't worked out the way I expected them to when using the parameter estimation tools in this package.gRain: I tried working with this a lot, but couldn't sort out how to generate the CPT values when one or more of the variables in my DAG was latent.
I also tried working out how to use rJags for a long time but ended up giving up. The tutorials I found for parameter estimation all assumed either:
- There are no latent variables
- We were trying to do learning on the structure, rather than just determine conditional probabilities given a structure.
I'd really like it if I could find a solid R package for tackling this problem, though a java/scala solution would work as well. A tutorial/example problem would be even better.
I have a vague impression that expectation maximization techniques can be used and even started trying to write code for the problem myself by extending the cluster graph stuff written here bayes-scala but quickly realized that this was a bigger project than I initially thought.
Bayes-Scala
I worked with Bayes scala some more and found a video from a coursera class defining cluster graphs http //www.youtube.com/watch?v=xwD_B31sElc
I'll walk through here quickly what I think I learned. First off, the full DAG I am dealing with is (http //i.imgur.com/tnVPFGM.png) (still not enough reputation for images or more links)
Most of the language dealing with cluster graphs refers to factors in factor graphs, so I tried to translate this DAG into a factor graph (http //i.imgur.com/V54XdTV.png). I'm not certain how this works, but I think this factor graph is a valid transformation from the DAG.
Then a simple cluster graph I could make is called a bethe cluster graph (http //i.imgur.com/hHb7eQ7.png). The bethe cluster graph is guaranteed to satisfy the constraints of cluster graphs. Namely:
1. Family Preservation: For each factor $\Phi_k$ there must exist a cluster $C_i$ s.t $scope(\Phi_k) \subseteq C_i$
2. Running Intersection Property: for each pair of clusters $C_i,C_j$ and variable $X_i \in C_i \cap C_j$ there exists a unique path between $C_i,C_j$ such that all clusters along the path contain $X_i$
However, I think the bethe cluster graph loses the covariability conferred by the original DAG. So I tried to construct what I believe is a valid cluster graph that still maintains information about covariance between variables:
http //i.imgur.com/54gDJhl.png
I think its possible that I messed something up in this cluster graph, but I could use bayes scala to write this cluster graph (http //pastebin.com/82ErCSQT). After I do so the log likelihood's progress as one would imagine:
EM progress(iterNum, logLikelihood): 1, -2163.880428036244 EM progress(iterNum, logLikelihood): 2, -1817.7287344711604
and the marginal probabilities can be calculated. Unfortunately the marginal probabilities for the hidden nodes turn out to be flat after using GenericEMLearn:
marginal D: WrappedArray(0.25, 0.25, 0.25000000000000006, 0.25000000000000006) marginal J: WrappedArray(0.25000000000000006, 0.25, 0.25, 0.25)
Additionally an odd error occurs where if I set evidence in a LoopyBP more than once I start getting NA values back:
val loopyBP = LoopyBP(skinGraph)
loopyBP.calibrate()
(1 to 44).foreach((sidx) => {
val s1 = dataSet.samples(sidx)
(1 to 10).foreach((x) => {
loopyBP.setEvidence(x,s1(x))
})
loopyBP.marginal(11).getValues.foreach((x) => print(x + " "))
println()
})
actual value was 0 0.0 0.0 0.0 1.0 actual value was 0 NaN NaN NaN NaN
This problem actually occurs for the sprinkler example as well, so I must be using setEvidence incorrectly somehow.