Follow the very useful answers from Peter Flom, Wayne and many others. I have now started using R and it gives me a feeling of python :)
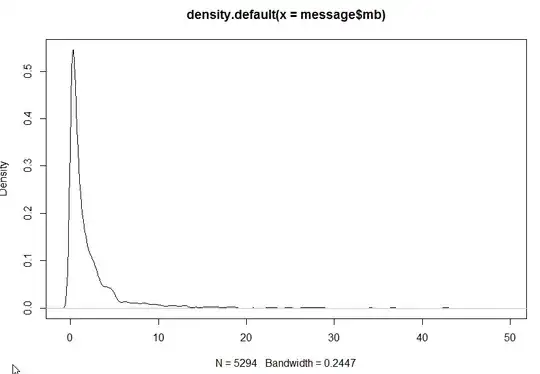
The results are below but I am not sure how should I go from here ? The density certain looks much better after log transformation. Can you please shed some light on how to do further analysis ?
Thanks a lot.
R - Results below:
plot (density (messages$length))
plot (density (log (messages$length)))
 summary (messages)
summary (messages)
> summary(message$mb)
Min. 1st Qu. Median Mean 3rd Qu. Max.
0.00665 0.32610 0.88450 2.08500 2.35000 49.13000
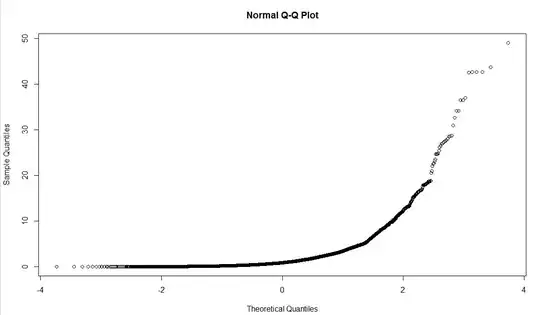
qqnorm (messages$length)
=====================================================================
EDIT: Thanks all all the answering !
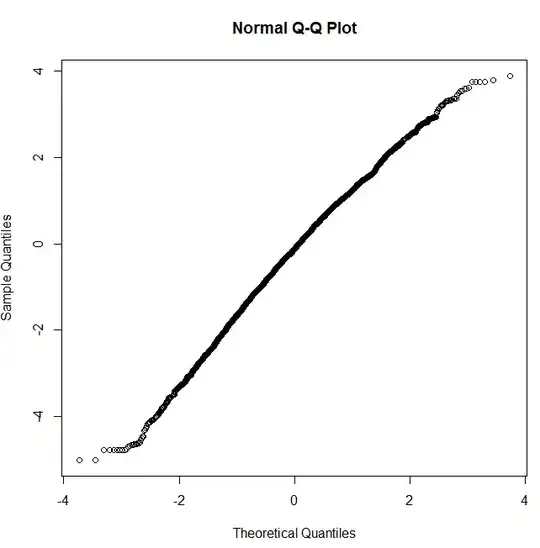
I have tried the qqnorm with log(x) and it looks like a straight line ! Does this mean my data is pretty much following a Log-normal distribution ?
qqnorm (log(messages$length)):
Also I have tried to fit my data with a log-normal and below is the result.
fitdistr(message$mb, densfun="log-normal") meanlog sdlog
-0.19019347 1.45795269 ( 0.02003787) ( 0.01416891)
Does this mean anything ?