I have fitted a certain ARMA model and got the following output:
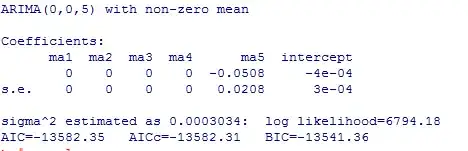
As you can see, some coeff were set to zero and the focus is on the ma5 coeff and the intercept. To be exactly, the ma5 coeff is -0.05077979 and the intercept is -0.00044509. Also the exact s.e. are 0.02078887 (ma5) and 0.00032599 (intercept). I want to calculate the p-values:
From the answer with 7 thumbs up on this thread (from mpiktas) I calculated it with:
(1-pnorm(abs(-0.05077979)/sqrt(0.02078887^2)))/2
which should give me the p-value for the ma5 coeff, and the output is
0.003645037
For the intercept I do
(1-pnorm(abs(-0.00044509)/sqrt(0.00032599^2)))/2
and get
0.04303588
Now I control this via coeff test of the lmtest package:
library(lmtest)
coeftest(myarmamodel)
and I get
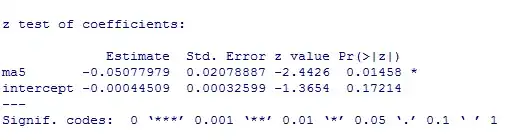
As you can see, the estimates and the s.e. are the same, but the p-value is different. My question is: Why? What is wrong? Is the p-value of the lmtest wrong or the solution in the other post?