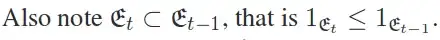I think you might do best to review multivariate regression (meaning no disrespect). There is a short tutorial at UVA's stats help page here: https://data.library.virginia.edu/getting-started-with-multivariate-multiple-regression/. They explain that multivariate regression is mostly the same as several univariate regressions, except that there are covariances between the betas for the different outcomes that need to be taken into account when testing the variables. In particular, they mention that the relevant plots are the same:
The same diagnostics we check for models with one predictor should be checked for these as well.
I will use their example to illustrate some data visualizations below (coded in R). I'll start with a scatterplot matrix of the data. GEN is binary, so I'll represent that with a different color and symbol. After fitting the model, if you try to run plot.lm() you'll get an error. However, it's easy enough to reproduce those plots manually. To plot a multiple regression model without interactions, you can pick a variable of interest and make a scatterplot with it and the response and draw the fitted function over it. Be sure to adjust the intercept by setting other variables at their means (see my answer to How to visualize a fitted multiple regression model?). You can also make scatterplots and qq-plots of the residuals (the latter lets you assess if their distribution is similar).
ami_data = read.table("http://static.lib.virginia.edu/statlab/materials/data/ami_data.DAT")
names(ami_data) = c("TOT","AMI","GEN","AMT","PR","DIAP","QRS")
summary(ami_data)
# output omitted
pairs(ami_data[,-3], col=ifelse(ami_data$GEN, "red", "black"),
pch=ifelse(ami_data$GEN, 3, 1))

mlm1 = lm(cbind(TOT, AMI) ~ GEN + AMT + PR + DIAP + QRS, data=ami_data)
summary(mlm1)
# output omitted
head(resid(mlm1))
# TOT AMI
# 1 132.82172 161.52769
# 2 -72.00392 -264.35329
# 3 -399.24769 -373.85244
# 4 -382.84730 -247.29456
# 5 -152.39129 15.78777
# 6 366.78644 217.13206
windows()
plot(mlm1)
# Error: 'plot.mlm' is not implemented yet
## reproducing R's plot.lm() for TOT
rst = rstandard(mlm1) # standardized residuals
windows()
layout(matrix(1:4, nrow=2, byrow=T))
# plot 1
plot(resid(mlm1)[,1]~fitted(mlm1)[,1],
main="Residuals vs Fitted for TOT", xlab="fitted", ylab="residuals")
lines(lowess(resid(mlm1)[,1]~fitted(mlm1)[,1]), col="red")
# plot 2
plot(sqrt(abs(rst[,1]))~fitted(mlm1)[,1],
main="Scale-Location plot for TOT", xlab="fitted", ylab="residuals")
lines(lowess(sqrt(abs(rst[,1]))~fitted(mlm1)[,1]), col="red")
# plot 3
qqnorm(rst[,1], main="qq-plot of residuals TOT")
qqline(rst[,1])
# plot 5
plot(rst[,1]~lm.influence(mlm1)$hat, xlab="Leverage", ylab="residuals",
main="Residuals vs Leverage for TOT")
lines(lowess(rst[,1]~lm.influence(mlm1)$hat), col="red")
windows()
layout(matrix(1:4, nrow=2, byrow=T))
# plot of model for TOT vs AMT
plot(TOT~AMT, ami_data, main="TOT vs AMT w/ data & model",
col=ifelse(ami_data$GEN, 2, 1), pch=ifelse(ami_data$GEN, 3, 1))
abline(coef(mlm1)[-3,1]%*%c(1, apply(ami_data[,c(3,5:7)], 2, mean)),
coef(mlm1)[3,1], col="blue")
# plot of model for AMI vs AMT
plot(AMI~AMT, ami_data, main="AMI vs AMT w/ data & model",
col=ifelse(ami_data$GEN, 2, 1), pch=ifelse(ami_data$GEN, 3, 1))
abline(coef(mlm1)[-3,2]%*%c(1, apply(ami_data[,c(3,5:7)], 2, mean)),
coef(mlm1)[3,2], col="blue")
# scatterplot of residuals
plot(resid(mlm1)[,1], resid(mlm1)[,2], xlab="Residuals for TOT",
ylab="Residuals for AMI", main="scatterplot of residuals")
# qq-plot of residuals
qqplot(resid(mlm1)[,1], resid(mlm1)[,2], xlab="Residuals for TOT",
ylab="Residuals for AMI", main="qq-plot for residuals")