As far as I understand, kernel density estimation does not make any assumptions on the moments of the underlying density, and just requires smoothness. The Cauchy density function is quite smooth. Even still, when I try to do KDE using density() in R for random draws from Cauchy distribution, I get incredibly inaccurate answers:
set.seed(1)
foo <- seq(-50, 50, length = 1e3)
plot(foo, dt(foo, df = 1), type = 'l')
lines(density(rt(1e3, df = 1)), col = "red")
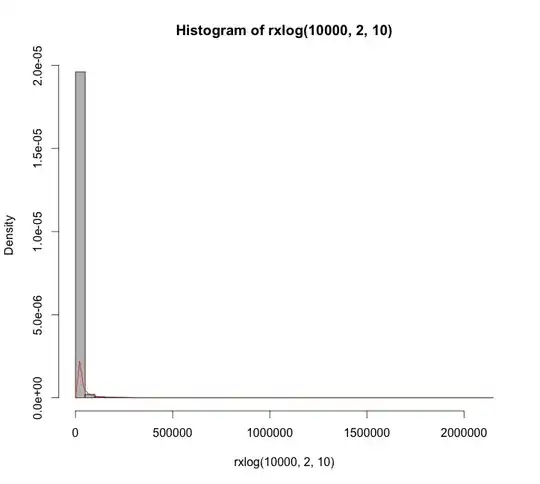
Repeat the above with different seeds or increasing the sample size can give further erratic estimates. The default kernel is Gaussian in R. Changing the kernel to any of the other options doesn't improve the output.
Question: What assumptions does Cauchy violate for KDEs? If it doesn't, then why do we see KDEs failing so miserably here?
Edit: @cdalitz has identified that the problem is where the kde is evaluating the density. The default is 3*bw*range(x), which for Cauchy can be quite large. Which means, by default density tries to estimate the KDE at 512 points sparsely distributed on the x-axis.
To test this, I change the from and to in the density estimation and see that if I run density twice with two sets of evaluating points, so the densities match:
set.seed(1)
samp <- rt(1e4, df = 1)
bd <- 10
den1 <- density(samp, from=-bd, to=bd, n=512)
den2 <- density(samp, from =-2*bd, to = 2*bd, n =512)
foo <- seq(-50, 50, length = 1e3)
plot(foo, dt(foo, df = 1), type = 'l')
lines(den2, col = "blue", type = "b")
lines(den1, col = "red", type = "b")
This produces the estimates below:
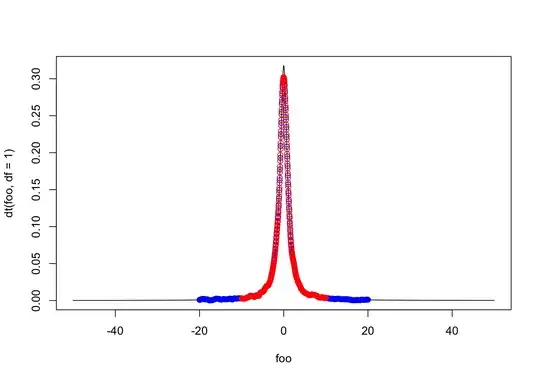
The quality here is much better than before. However, now if instead of 2*bd, I change this to 50*bd, I get that the density estimate even around 0 is very different!
set.seed(1)
samp <- rt(1e4, df = 1)
bd <- 10
den1 <- density(samp, from=-bd, to=bd, n=512)
den2 <- density(samp, from =-50*bd, to = 50*bd, n =512)
foo <- seq(-50, 50, length = 1e3)
plot(foo, dt(foo, df = 1), type = 'l', ylim = c(0,.7))
lines(den2, col = "blue", type = "b")
lines(den1, col = "red", type = "b")
How does evaluating the density at sparse points change the density evaluation process around $x = 0$ (the bandwidth chosen is the same for both den1 and den2)? The KD estimate at any point $x$ is
$$
\hat{f}(x) = \dfrac{1}{nh} \sum_{t=1}^{n} K\left( \dfrac{x - x_i}{h}\right)\,.
$$
The density estimate shouldn't change at a given value of $x = a_1$ if the density is also being evaluated at other points. What am I missing here?