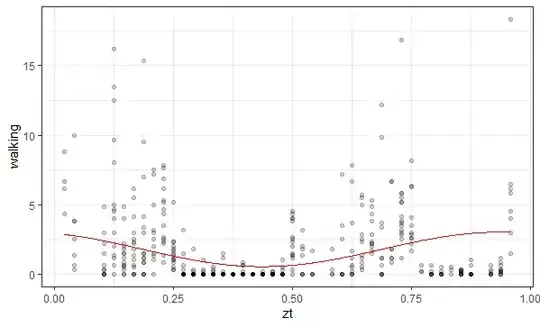
I'm trying to fit a sine wave over some activity data, just like this post. I've managed to get a reasonable looking graph for one condition:
However, when I plot the other condition the wave looks incredibly weird and not what it should look like --- it should be nice and smooth like the first image. Also, it should have two peaks and one trough like the first image.
What's going wrong here? Is there anything I can do to fix this? The second condition has the same 48 hours, etc.
My data has hourly measurements across a 48 hour period (two consecutive days, so there should be one peak and one trough/day). Code was adapted from here.
b$zt <- b$zt/48
lmfit <- lm(data = b,
walking ~ sin(2*pi*b$zt) + cos(2*pi*b$zt))
b0 <- coef(lmfit)[1]
alpha <- coef(lmfit)[2]
beta <- coef(lmfit)[3]
pframe <- data.frame(zt=seq(min(b$zt),max(b$zt),length=612))
pframe$walking <- predict(lmfit,newdata=pframe)
library(ggplot2); theme_set(theme_bw())
ggplot(b,aes(zt,walking))+
geom_point(alpha=0.2) + geom_line(data=pframe,colour="red")
Here are the relevant parts from the dataset:
dput(new$zt)
c(6L, 6L, 6L, 6L, 7L, 7L, 7L, 7L, 8L, 8L, 8L, 8L, 9L, 9L, 9L,
9L, 11L, 11L, 11L, 11L, 12L, 12L, 12L, 12L, 15L, 15L, 15L, 15L,
17L, 17L, 17L, 17L, 23L, 23L, 23L, 23L, 24L, 24L, 24L, 24L, 1L,
1L, 1L, 1L, 5L, 5L, 5L, 5L, 6L, 6L, 6L, 6L, 7L, 7L, 7L, 7L, 8L,
8L, 8L, 8L, 9L, 9L, 9L, 9L, 10L, 10L, 10L, 10L, 11L, 11L, 11L,
11L, 12L, 12L, 12L, 12L, 13L, 13L, 13L, 13L, 14L, 14L, 14L, 14L,
15L, 15L, 15L, 15L, 17L, 17L, 17L, 17L, 19L, 19L, 19L, 19L, 24L,
24L, 24L, 24L, 5L, 5L, 5L, 5L, 6L, 6L, 6L, 6L, 7L, 7L, 7L, 7L,
8L, 8L, 8L, 8L, 9L, 9L, 9L, 9L, 10L, 10L, 10L, 10L, 11L, 11L,
11L, 11L, 12L, 12L, 12L, 12L, 22L, 22L, 22L, 22L, 24L, 24L, 24L,
24L, 2L, 2L, 2L, 2L, 5L, 5L, 5L, 6L, 6L, 6L, 7L, 7L, 7L, 8L,
8L, 8L, 9L, 9L, 9L, 10L, 10L, 10L, 11L, 11L, 11L, 12L, 12L, 12L,
14L, 14L, 14L, 15L, 15L, 15L, 16L, 16L, 16L, 17L, 17L, 17L, 18L,
18L, 18L, 19L, 19L, 19L, 21L, 21L, 21L, 22L, 22L, 22L, 23L, 23L,
23L, 24L, 24L, 24L, 2L, 2L, 2L, 31L, 31L, 31L, 31L, 32L, 32L,
32L, 32L, 33L, 33L, 33L, 33L, 35L, 35L, 35L, 35L, 36L, 36L, 36L,
36L, 39L, 39L, 39L, 39L, 41L, 41L, 41L, 41L, 42L, 42L, 42L, 42L,
44L, 44L, 44L, 44L, 45L, 45L, 45L, 45L, 46L, 46L, 46L, 46L, 25L,
25L, 25L, 25L, 26L, 26L, 26L, 26L, 28L, 28L, 28L, 28L, 30L, 30L,
30L, 30L, 35L, 35L, 35L, 35L, 29L, 29L, 29L, 29L, 30L, 30L, 30L,
30L, 31L, 31L, 31L, 31L, 32L, 32L, 32L, 32L, 33L, 33L, 33L, 33L,
34L, 34L, 34L, 34L, 35L, 35L, 35L, 35L, 36L, 36L, 36L, 36L, 38L,
38L, 38L, 38L, 41L, 41L, 41L, 41L, 42L, 42L, 42L, 42L, 45L, 45L,
45L, 45L, 46L, 46L, 46L, 46L, 29L, 29L, 29L, 30L, 30L, 30L, 31L,
31L, 31L, 32L, 32L, 32L, 33L, 33L, 33L, 34L, 34L, 34L, 35L, 35L,
35L, 36L, 36L, 36L, 37L, 37L, 37L, 38L, 38L, 38L, 39L, 39L, 39L,
40L, 40L, 40L, 41L, 41L, 41L, 42L, 42L, 42L, 44L, 44L, 44L, 45L,
45L, 45L, 25L, 25L, 25L, 13L, 13L, 13L, 13L, 14L, 14L, 14L, 14L,
16L, 16L, 16L, 16L, 18L, 18L, 18L, 18L, 19L, 19L, 19L, 19L, 20L,
20L, 20L, 20L, 21L, 21L, 21L, 21L, 22L, 22L, 22L, 22L, 13L, 13L,
13L, 13L, 14L, 14L, 14L, 14L, 15L, 15L, 15L, 15L, 17L, 17L, 17L,
17L, 18L, 18L, 18L, 18L, 21L, 21L, 21L, 21L, 16L, 16L, 16L, 16L,
18L, 18L, 18L, 18L, 20L, 20L, 20L, 20L, 21L, 21L, 21L, 21L, 22L,
22L, 22L, 22L, 23L, 23L, 23L, 23L, 13L, 13L, 13L, 13L, 14L, 14L,
14L, 14L, 15L, 15L, 15L, 15L, 16L, 16L, 16L, 16L, 17L, 17L, 17L,
17L, 18L, 18L, 18L, 18L, 19L, 19L, 19L, 19L, 20L, 20L, 20L, 20L,
21L, 21L, 21L, 21L, 14L, 14L, 14L, 14L, 16L, 16L, 16L, 16L, 17L,
17L, 17L, 17L, 18L, 18L, 18L, 18L, 21L, 21L, 21L, 21L, 22L, 22L,
22L, 22L, 13L, 13L, 13L, 20L, 20L, 20L, 13L, 13L, 13L, 14L, 14L,
14L, 17L, 17L, 17L, 21L, 21L, 21L)
> dput(new$walking)
c(8, 1.166666667, 4.666666667, 2, 4.833333333, 2, 1.833333333,
4, 2.5, 1.333333333, 4.166666667, 1.833333333, 7, 1.166666667,
9.5, 1.166666667, 6.833333333, 5.166666667, 4.333333333, 2.333333333,
5.166666667, 2.333333333, 2, 1.833333333, 0, 0, 0, 0, 0, 0, 1.5,
0, 0, 0, 1, 0.333333333, 0.333333333, 1.5, 2.5, 2.333333333,
4.333333333, 8.833333333, 6.666666667, 6.166666667, 2.166666667,
0.333333333, 0, 0, 9.666666667, 12.5, 13.5, 16.16666667, 0, 2.166666667,
0.666666667, 0.5, 0, 2.333333333, 0, 0, 0, 1.5, 3, 0.833333333,
3, 7.5, 5, 3.166666667, 7.666666667, 7.833333333, 2.666666667,
2.5, 2.333333333, 1, 1.666666667, 0.333333333, 1.5, 0.666666667,
0.333333333, 3.166666667, 1.833333333, 0, 0, 0, 0, 1, 0, 0, 0,
1.166666667, 0, 0, 0, 0.833333333, 0.666666667, 0.666666667,
0, 4, 4.5, 3.833333333, 0, 3, 0.333333333, 0, 4.5, 1.333333333,
5, 3, 0, 0.666666667, 0.333333333, 1.5, 3, 5.5, 1, 6.166666667,
1.5, 3.333333333, 15.33333333, 4.166666667, 2, 1.333333333, 7.166666667,
1.5, 1.333333333, 5.333333333, 4.666666667, 6.166666667, 0.333333333,
1.166666667, 0.833333333, 1.5, 0, 0, 0, 0.166666667, 1.166666667,
2, 1.333333333, 4.333333333, 3.833333333, 1.333333333, 0.333333333,
0.666666667, 1.5, 4.833333333, 0.666666667, 0.666666667, 0, 4,
1.5, 0.833333333, 1.166666667, 2.166666667, 0.5, 0.666666667,
0, 0, 0.833333333, 2.5, 1, 0, 0.833333333, 3.5, 2.166666667,
1, 1.333333333, 0.5, 0.333333333, 0.166666667, 0, 0, 0.166666667,
0.333333333, 0.333333333, 0.166666667, 0.5, 0, 0.166666667, 0,
0, 0, 0, 0, 0.166666667, 0, 0, 0, 0.166666667, 0, 0.166666667,
0, 0, 0, 0.5, 1.333333333, 4, 2.666666667, 3.833333333, 2.5,
10, 1.166666667, 5, 2.5, 2.333333333, 3.833333333, 4.833333333,
2, 2.833333333, 0, 0.666666667, 0, 0, 3, 3.5, 5.166666667, 3.166666667,
3.5, 2.833333333, 2.166666667, 8.166666667, 0.166666667, 0.5,
0, 0, 0.333333333, 0.166666667, 0.333333333, 0, 0, 0.666666667,
0, 0, 0.166666667, 0.333333333, 0.666666667, 0, 0.166666667,
0.166666667, 1.166666667, 0.666666667, 3, 6.5, 4, 6.166666667,
0, 3.166666667, 1.333333333, 2, 0, 0, 1.666666667, 0, 1.833333333,
0, 0.333333333, 0, 0, 0.166666667, 0.5, 0, 3.666666667, 1.166666667,
3.833333333, 2.5, 0, 0, 0, 0, 1.666666667, 1.166666667, 0.666666667,
0, 0.833333333, 2.833333333, 1.333333333, 0.5, 2.166666667, 1.5,
1.666666667, 4.333333333, 12.16666667, 9.833333333, 3.666666667,
3.333333333, 1.166666667, 6.666666667, 1.833333333, 1.166666667,
5.5, 4.166666667, 5.833333333, 16.83333333, 2.602230483, 6.319702602,
1.672862454, 4.089219331, 0, 0.666666667, 0, 0, 0.166666667,
0.166666667, 0, 0, 0, 0, 0, 1, 0, 0.166666667, 0.333333333, 0.833333333,
18.33333333, 5.833333333, 1.5, 4.5, 3.5, 7.166666667, 2.5, 7.833333333,
3.666666667, 6.666666667, 0.333333333, 5.666666667, 5.5, 2.166666667,
5.333333333, 2.333333333, 1.166666667, 1.166666667, 0, 0.833333333,
6.666666667, 1.833333333, 2.833333333, 5.833333333, 5.166666667,
2.941176471, 2.573529412, 6.25, 0.166666667, 0.666666667, 0.166666667,
0, 0, 0.166666667, 0, 0, 0, 0.5, 0, 0.333333333, 0, 0, 0.333333333,
0, 0, 0, 0, 0, 0, 0, 0.333333333, 0, 0, 0, 1.833333333, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0)