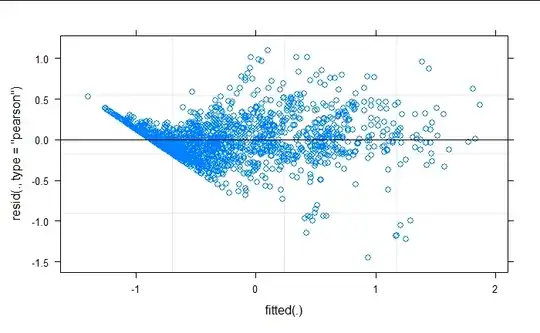
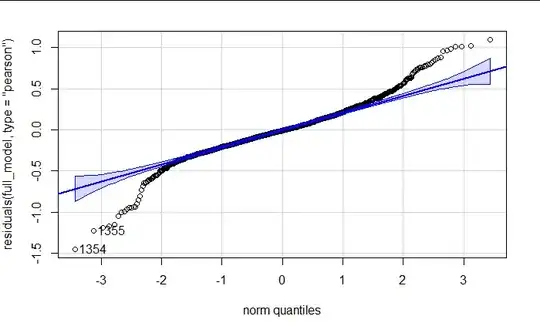
I am attempting to model an experiment which was measuring the production of co2 and ch4 at varying temperatures. The experiment has two underlying treatments: substrate with three levels and climate with three levels. I have tried fitting both a gamma glmer with log link and a log transformed lmer. The glmer fails to converge even with optimization, the lmer has no convergence issues is struggling with prediction accuracy at the lower and higher bounds.
#non-converging model
full_model_Gamma <- glmer(co2_accumulation ~ substrate #+ climate
+ ch_temp
+ treatment
+ waterbath
+ ch_temp:substrate
+ ch_temp:treatment
+ treatment:substrate
+ (1|site)
, family = Gamma(link = 'log')
, data = Oxymax#[waterbath == "Old",]
, na.action = "na.fail"
, control=glmerControl(optimizer="nloptwrap",optCtrl=list(algotithm = "NLOPT_LN_NELDERMEAD")))
#converging model
full_model <- lmer(log(co2_accumulation_s+1) ~ substrate + ch_temp_s
#+ climate removed from drop1
+ treatment
+ waterbath
+ ch_temp_s:substrate
+ ch_temp_s:treatment
+ treatment:substrate # SIR treatments could increase the CO2 rate of calc sites more than period ones due to more microbes
#+ substrate:climate
#+ (1+treatment|site)
+ (1|site)
, data = Oxymax#[waterbath == "Old",]
, na.action = "na.fail"
, control=lmerControl(optimizer="bobyqa",optCtrl=list(maxfun=2e5))
)
qqPlot(residuals(full_model, type = "pearson"))
plot(residuals(full_model_Gamma, type = "pearson") ~ factor(Oxymax$substrate))
plot(residuals(full_model_Gamma, type = "pearson") ~ factor(Oxymax$climate))
plot(residuals(full_model_Gamma, type = "pearson") ~ Oxymax$ch_temp) #within this you see potential interaction between ch temp and
Any help would be greatly appreciated.