I have a panel dataset which I have fit a fixed-effects model to using plm() in R:
# Sample panel data
ID, year, progenyMean, damMean
1, 1, 70, 69
1, 2, 68, 69
1, 3, 72, 72
1, 4, 69, 68
2, 1, 76, 75
2, 2, 73, 80
3, 1, 72, 74
3, 2, 75, 67
3, 3, 71, 69
# Fixed Effects Model in plm
fixed <- plm(progenyMean ~ damMean, data, model= "within", index = c("ID","year"))
I have plotted progenyMean vs damMean with the fixed effects regression line in blue:
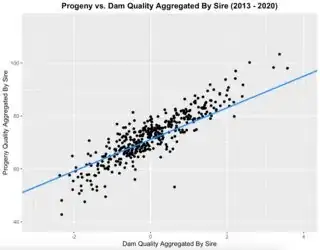
There are several data points for every unique ID, so it's possible for an ID to have points both above and below the regression line.
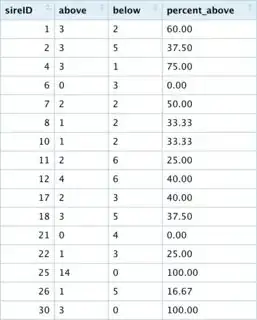
I have identified the data points above/below the regression line and I want to rank the ID's according to them being above the regression line. I created a table where each row is a unique ID and there are two columns, above and below which count the number of data points above/below the regression line for that ID. The last column percent_above is the percentage of each ID's data points that are above the line.
My question is thus: Is there a way to rank these ID's in terms of being above the regression line? There are 2 possibilities I have considered:
- Rank ID's according to the proportion of their data points above the regression line. The problem here is that certain ID's with only 3 observations total (all above the line) will rank higher than an ID with 13 observations above the line and 1 observation below the line.
- Rank ID's according to the number of observations above the line. The problem here is that an ID with 7 points above and 4 below will rank higher than an ID with all 5 data points above the line.
If there is another way that I have not considered please let me know.