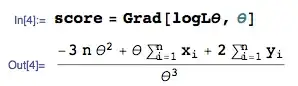This is my data and I'd like to do REML analysis to see variance components of random factor.
Plant<- rep(c("P1","P2","P3","P4"), each=9)
Leaves<- rep(rep(c("L1","L2","L3"), each=3),4)
Rep<-rep(c(1,2,3),12)
Ca<- c(3.280, 3.090, 3.185, 3.520, 3.600, 3.560, 2.880, 2.800, 2.840, 2.460, 2.440,
2.450, 1.870, 1.800, 1.835, 2.190, 2.100, 2.145, 2.770, 2.660, 2.715, 3.740,
3.440, 3.590, 2.550, 2.700, 2.625, 3.780, 3.870, 3.825, 4.070, 4.200, 4.135,
3.310, 3.400, 3.355)
tomato<- data.frame(Plant,Leaves,Rep,Ca)
and this is my code
library(lme4)
lmer <- lmer(Ca ~ (1|Plant)+ (1|Plant:Leaves), REML=TRUE, data=tomato)
summary(lmer)
I assumed that leaves are nested to plant. So I code as (1|Plant:Leaves)
This result indicates that Plant and leaves account for the most variability in data, while replicates are minor, doesn't it?
Then, I want to know plant and leave (nested to plant) are significant or not. Where can I find the p-value? or can I add more code to check p-value?
Or in REML, only variance component is possible for us to check?
If so, if I choose plant as a fixed factor like below
lmer <- lmer(Ca ~ Plant+ (1|Plant:Leaves), REML=TRUE, data=tomato)
summary(lmer)
At least, in this mixed model, p-value for plant (= fixed factor) should be presented? isn't it? But, I can't still find it.
Could you guide me how to analyze the result of REML?