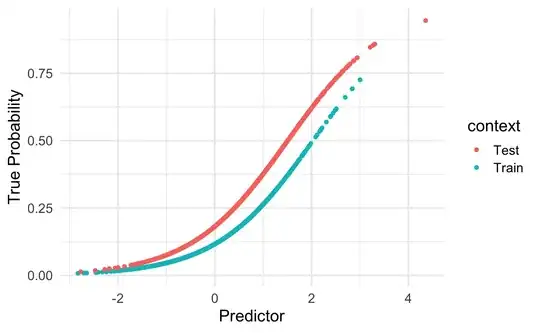
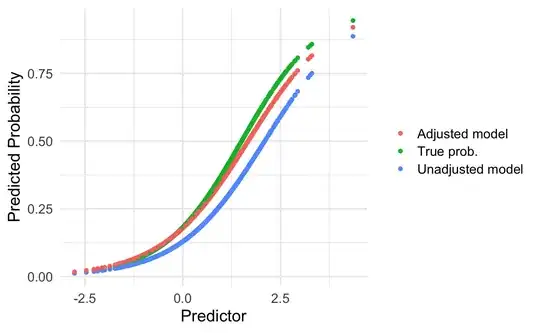
I wonder if thresholds for logistic regression models are prevalence-specific. I assume that they are, however, I am not sure about the basic statistical principles behind it and how to deal with the implications for clinical practice.
Example:
A hospital wants do deploy a logistic regression model to predict lymph node metastasis in prostate cancer patients. The model is recommended by a specialist society and widely accepted in the medical community.
For model development, a research group used a large dataset where the prevalence of lymph node metastasis was low (15%). They used a lab value (PSA) and age as predictors. After external validation with data from hospitals with similar prevalence (15 %), decision curve analysis and discussing the benefits and harms of the treatment the specialist society found a threshold probability of ≥0.10 appropriate regarding decision if a patient needs specific surgery (medically reasonable amount of true positive and false positive results).
Now the hospital is deploying the model in their surgery consultation-hour (expected prevalence of Patients with lymph node metastasis = 30%).
Questions:
- Can they deploy the same threshold probability if they want to have similar true positive and false positive results?
- If not, how should the model and/or threshold probability be adjusted (to get similar true positive and false positive results)?
What I already found on this topic:
An interresting blog about prevalence and probability, however, it does not answer my question regarding thresholds.
The Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD) statement (W17):
In general, models will be more generalizable when the case mix of the new population is within the case mix range of the development population (186). However, as we describe under item 10e (see also Box C and Table 3), one may adjust or update a previously developed prediction model that is applied in another setting to the local circumstances of the new setting to improve the model transportability.
from W17 Table 3:
Updating Method: Adjustment of the intercept(baseline risk/hazard)
Reason for Updating: Difference in the outcome frequency (prevalence or incidence) between development and validation sample
Reproducible Example in R:
#library
library(tidyverse)
library(rmda)
# train data (prevalence= 15%)
train <- tibble(id=1:1000,
class=c(rep(1,150),rep(0,850)))
set.seed(1)
train %>%
group_by(id) %>%
mutate(
PSA=case_when(class==1 ~ runif(1,1,100),TRUE ~ runif(1,1,40)),
Age=case_when(class==1 ~ runif(1,30,80),TRUE ~runif(1,30,60))) -> d.train
# test data same prevalence (15%)
test <- tibble(id=1:1000,
class=c(rep(1,150),rep(0,850)))
set.seed(23)
test %>%
group_by(id) %>%
mutate(
PSA=case_when(class==1 ~ runif(1,1,100),TRUE ~ runif(1,1,50)),
Age=case_when(class==1 ~ runif(1,30,80),TRUE ~runif(1,25,60))) -> d.test_same_prev
# test data high prevalence (30%)
test1 <- tibble(id=1:1000,
class=c(rep(1,350),rep(0,650)))
set.seed(123)
test1 %>%
group_by(id) %>%
mutate(
PSA=case_when(class==1 ~ runif(1,1,100),TRUE ~ runif(1,1,50)),
Age=case_when(class==1 ~ runif(1,30,80),TRUE ~runif(1,25,60))) -> d.test_higher_prev
# train logistic regression model
glm(class ~ Age+PSA, data=d.train,family = binomial) -> model
# make predictions in cohort with same prevalence
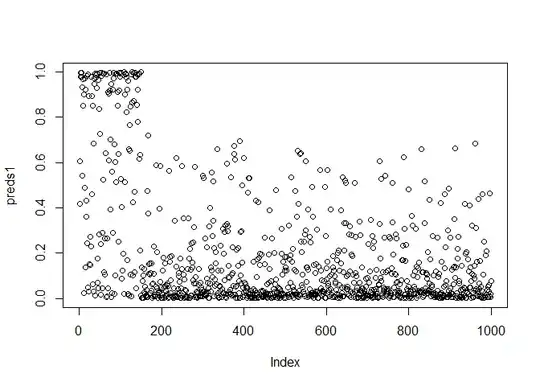
predict(model,d.test_same_prev, type="response") -> preds1
plot(preds1)
# make predictions in cohort with high prevalence
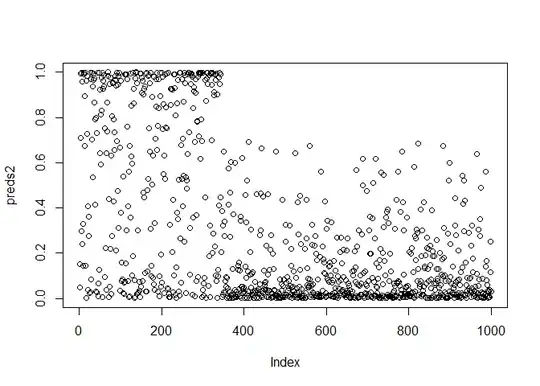
predict(model,d.test_higher_prev, type="response") -> preds2
plot(preds2)
# decision curve analysis same prevalence
d.dca.same <- data.frame(reference=d.test_same_prev$class,predictor=preds1)
dca.same <-decision_curve(reference ~predictor,d.dca.same,fitted.risk=TRUE, bootstraps = 10)
plot_decision_curve(dca.same,confidence.intervals=FALSE)
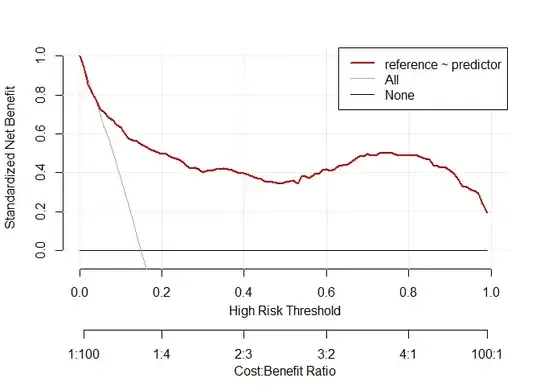
# decision curve analysis high prevalence
d.dca.high <- data.frame(reference=d.test_higher_prev$class,predictor=preds2)
dca.high <-decision_curve(reference ~predictor,d.dca.high,fitted.risk=TRUE, bootstraps = 10)
plot_decision_curve(dca.high,confidence.intervals=FALSE)
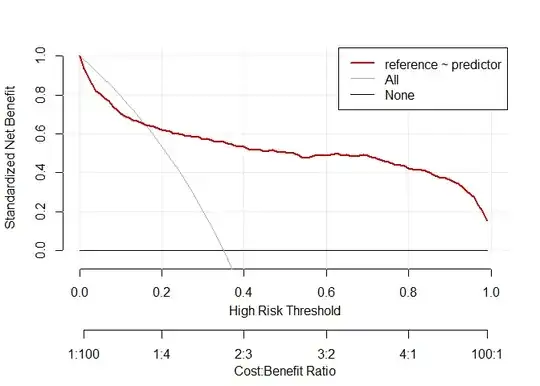
Created on 2021-08-08 by the reprex package (v2.0.0)