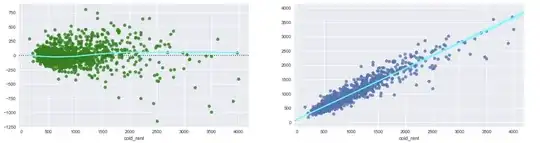
I am a bit confused about how to approximate the equation from a nonlinear model constructed in brms, and was hoping someone could explain it to me.
Say I have the below model:
Family: lognormal
Links: mu = identity; sigma = identity
Formula: weight.g ~ b1^b2
b1 ~ 1 + dia.1.cm
b2 ~ 1
Data: CaesCombBot.data.wfrag (Number of observations: 2619)
Samples: 4 chains, each with iter = 5000; warmup = 2000; thin = 5;
total post-warmup samples = 2400
Population-Level Effects:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
b1_Intercept 6.61 0.81 5.08 8.24 1.00 1741 1787
b1_dia.1.cm 0.98 0.42 0.42 2.07 1.00 1738 1974
b2_Intercept 0.57 0.06 0.46 0.70 1.00 1727 1942
Family Specific Parameters:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sigma 0.56 0.01 0.54 0.57 1.00 2156 2105
Samples were drawn using sampling(NUTS). For each parameter, Bulk_ESS
and Tail_ESS are effective sample size measures, and Rhat is the potential
scale reduction factor on split chains (at convergence, Rhat = 1).
I'm assuming that the supplied formula represents something along the lines of (int + ax)^b - but when I try to place the estimates back into the equation, they appear to do a poor job of approximating the model displayed.
For example:
curve((6.61 + 0.98*x)^0.57, from = 0, to = 50)
Just doing a quick comparison: x value of 40 (6.61 + 0.98*40)^0.57 = exp(8.846) [modelled in lognormal dist] = 6946.547
Compared to x value of 40 corresponding to 5256.29 according to add_predictions for model
I understand they visually appear different as one is on the log scale while one is on the response scale, but shouldn't the values roughly match up at least?
I feel as though I am probably missing something very obvious and fundamental, would be very grateful for any help or insight.