I'm having a hard time trying to understand the differences between these two models and why the first one shows correlation (p-value < 0,05) but the other one doesn´t (p-value > 0,05).
I would be very grateful if someone could help me, because I have to submit my thesis soon.
In this study we want to assess if the human balance improves after training.
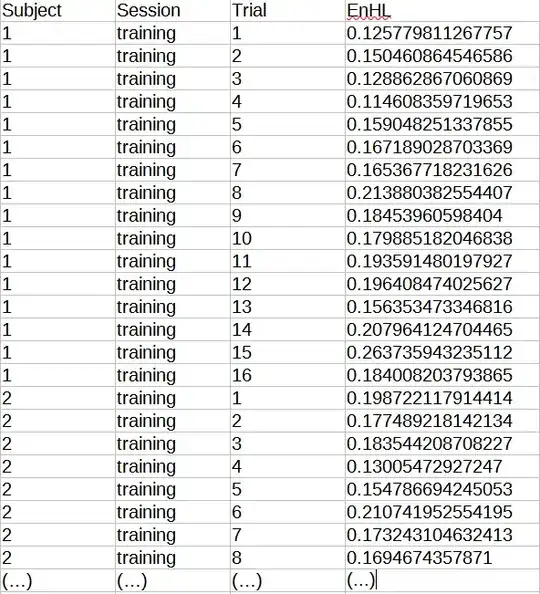
We had 40 subjects. Our dependent variable (EnHL, the one that gives us the information about balance) was measured three times over one week for each subject (training, 1 day later and 7 days later). In the first day (training) each subject did 16 trials, 1 day later they did 4 trials and 7 days later they did 16 trials.
My table looks like this: (2881 rows and 4 columns)
Here's a plot of all the trials within the sessions for each subject
My question is: what model would be more accurate?
gamma <- glmmTMB(EnHL ~ Session + Trial + (1 | Subject) , data = original, family = "Gamma" (link = log))
glmmTMB:::Anova.glmmTMB(gamma,contrasts=list(Session=contr.sum, Trial=contr.sum), type = 3)
Response: EnHL
Chisq Df Pr(>Chisq)
(Intercept) 2118.0801 1 < 2.2e-16 ***
Session 11.9676 2 0.002519 **
Trial 6.1979 1 0.012790 *
summary(gamma)
Family: Gamma ( log )
Formula: EnHL ~ Session + Trial + (1 | Subject)
Data: original
AIC BIC logLik deviance df.resid
-4738.3 -4706.9 2375.2 -4750.3 1391
Random effects:
Conditional model:
Groups Name Variance Std.Dev.
Subject (Intercept) 0.03568 0.1889
Number of obs: 1397, groups: Subject, 40
Dispersion estimate for Gamma family (sigma^2): 0.0435
Conditional model:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -1.579152 0.034390 -45.92 < 2e-16 ***
Session7days 0.046246 0.020271 2.28 0.02253 *
Sessiontraining 0.012743 0.020226 0.63 0.52868
Trial -0.003473 0.001271 -2.73 0.00627 **
here the p-value for the trials and for the sessions is less than 0,05 (there's a relationship between sessions (after 7 days) and EnHL and between trials and EnHL)
or
gammacrs <- glmmTMB(EnHL ~ Session + Trial + (Trial + Session | Subject), data = original, family = "Gamma" (link = log))
where I´m assuming that the sessions are allowed to vary between the subjects. (If this wasn't the case nothing should change, right?)
glmmTMB:::Anova.glmmTMB(gammacrs,contrasts=list(Session=contr.sum, Trial=contr.sum), type = 3)
Response: EnHL
Chisq Df Pr(>Chisq)
(Intercept) 1638.3203 1 <2e-16 ***
Session 4.2577 2 0.1190
Trial 2.6027 1 0.1067
summary(gammacrs)
Family: Gamma ( log )
Formula: EnHL ~ Session + Trial + (Trial + Session | Subject)
Data: original
AIC BIC logLik deviance df.resid
-4828.5 -4749.9 2429.3 -4858.5 1382
Random effects:
Conditional model:
Groups Name Variance Std.Dev. Corr
Subject (Intercept) 0.0513038 0.226503
Trial 0.0000902 0.009497 0.17
Session7days 0.0126242 0.112357 -0.69 -0.31
Sessiontraining 0.0250126 0.158154 -0.46 -0.51 0.59
Number of obs: 1397, groups: Subject, 40
Dispersion estimate for Gamma family (sigma^2): 0.0369
Conditional model:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -1.583330 0.039118 -40.48 <2e-16 ***
Session7days 0.044660 0.025793 1.73 0.0834 .
Sessiontraining 0.011696 0.031197 0.37 0.7077
Trial -0.003073 0.001905 -1.61 0.1067
With this model there are no correlations between sessions and EnHL or between trials and EnHL anymore (p-value > 0,05).
The function anova(gamma, gammacrs, test = "Chisq") tells us that the second model is better.
Models:
gamma: EnHL ~ Session + Trial + (1 | Subject), zi=~0, disp=~1
gammacrs: EnHL ~ Session + Trial + (Trial + Session | Subject), zi=~0, disp=~1
Df AIC BIC logLik deviance Chisq Chi Df Pr(>Chisq)
gamma 6 -4738.3 -4706.9 2375.2 -4750.3
gammacrs 15 -4828.5 -4749.9 2429.3 -4858.5 108.22 9 < 2.2e-16 ***
My interpretation of the results is: The sessions and the trials are different for every subject. Some of them improve but others don't (the differences between subjects are important). What do you think?
Edit:
I've extracted all the random effects.
Note 1 : Intercept = Training ; Session1 = 1day ; Session2 = 7days
$Subject
(Intercept) Session1 Session2 Trial
1 -1.713896 0.0965891767 -0.011518628 0.0067663851
2 -1.561364 0.0840375097 0.080187190 0.0137080183
3 -1.275755 0.0004848607 -0.042586717 0.0028534510
4 -1.278252 -0.0536642568 -0.020647458 -0.0122991059
5 -1.465870 0.0181134542 0.008405354 -0.0002592055
6 -1.610363 -0.0988696152 0.012303906 -0.0069000874
7 -1.464979 0.0925183339 0.015557069 0.0010255370
8 -1.804658 -0.1398576157 0.102382968 -0.0060396435
9 -1.801174 -0.0529431098 0.024780565 -0.0042638591
10 -1.421477 -0.0921801843 0.065533093 -0.0041786959
11 -1.480386 0.0144059690 0.057358732 -0.0047655723
12 -1.386061 0.0902745005 0.046153060 -0.0027204657
13 -1.668770 0.0219507766 0.084439469 -0.0037810769
14 -1.716096 0.0488681197 0.176934080 0.0043043320
15 -1.784928 -0.1327412005 0.085074360 -0.0086768685
16 -1.532577 0.0272251821 -0.038856126 -0.0077024692
17 -1.725039 -0.0909731768 -0.015877398 -0.0113995813
18 -1.322610 0.0287808148 -0.040762983 0.0031696644
19 -1.411264 0.0477764606 -0.007162943 0.0040797905
20 -1.429356 -0.0084597510 -0.033387436 -0.0047225067
21 -1.555885 0.0571250855 0.029741363 0.0072087558
22 -1.630751 -0.1443444132 -0.036059771 -0.0261584712
23 -1.796652 -0.0375694398 0.058203264 -0.0109808637
24 -1.469535 -0.0100575168 0.011208402 -0.0113491972
25 -1.973101 -0.0727788418 0.098841831 -0.0080184843
26 -1.675099 -0.0494589192 0.059443376 0.0071001076
27 -1.245882 0.0011772501 -0.019320935 -0.0034227885
28 -1.558100 -0.0239727664 0.062121576 -0.0057094588
29 -1.474227 -0.0059313380 0.091154895 0.0077893881
30 -1.404088 -0.0543213259 -0.023103316 -0.0004199302
31 -1.877645 -0.0784904402 0.070055279 0.0005748268
32 -1.409313 -0.0320830203 -0.038264689 -0.0014453813
33 -1.614586 -0.0427011119 -0.008117112 -0.0093748427
34 -1.452076 0.0320788297 0.039076646 -0.0182604601
35 -1.710346 -0.0326692685 0.002930763 0.0042231651
36 -1.836628 -0.0838170332 0.058713821 -0.0084572632
37 -1.623402 -0.0400794014 0.036481544 0.0029131856
38 -1.505535 0.0388458313 -0.007707702 0.0026484699
39 -1.292290 -0.0282789746 -0.008594992 0.0016093783
40 -1.681294 -0.0675433733 0.022157397 -0.0121784692
and ploted them
Note 2 : The plot doesn't exactly match the "coef" table. Some red points are positive in the table. Is this because of the standard error?