I would like to compare two methods on qualitative and paired data.
To summarize my study for you (it's for an internship), I have activity values that were measured on 13 sites, for 12 species of bats (some species are not found on all sites).
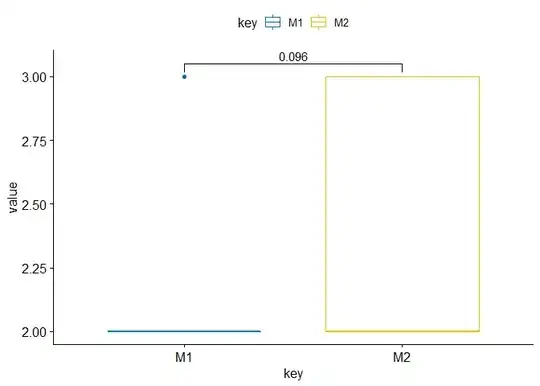
These activity values are then organized according to 4 levels: weak, medium, strong and very strong, by two different methods. I would like to know if these methods give significantly different activity levels, and if so, which method gives higher activity levels in general?
Then I want to compare these methods on smaller samples, such as groups of species or just by specie (so with only a few samples). I think method 1 will sometimes give higher levels than method 2 depending on the species group formed, and vice versa.
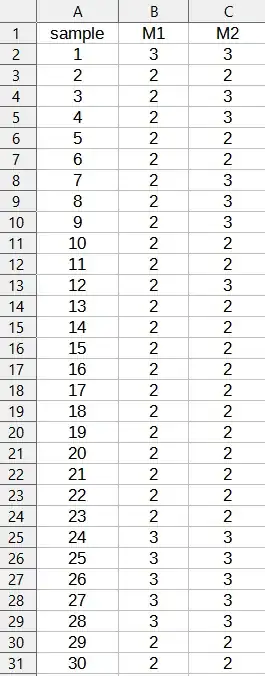
Here is an example from my data: (I have 132 samples in total)
On M1 and M2 you have the activity levels : 1 = weak, 2 = medium, 3 = strong and 4 = very strong
What test could I perform on R to compare these methods and tell which one produces higher activity levels? Also, I would like to present my results in a fairly visual way, with graphics. What is possible to do?
I apologize if there are any mistakes, english is not my native language. Also I'm new to this website !