I've looked through multiple previous questions on this topic and I have made some progress understanding continuous/continuous interactions with a GAM, but I still need some help understanding my results and making sure my modeling approach is statistically sound. I'm still very new to modeling with GAMs. I'm using mgcv in R.
I have a principle (nonlinear) relationship of y~s(x)+blocking_var. Now, we're interested in whether/how a number of continuous covariates qualitatively change that relationship. We are not interested in the main effects of those covariates as they will not be real-life causal predictors of y.
All covariates we're interested in are continuous. There may be some collinearity and we suspect nonlinear interactions. Our main predictor and the other covariates are on different units/scales, e.g. normalized counts:julian day, normalized counts:percentage of counts of group A etc.
For an interaction with one additional covariate, I understand I can do y~te(x,cov1) + blocking_var. But, how do I incorporate multiple factors?
My questions are:
Do I actually need to use te() instead of s() as in
y~s(x,cov1) + blocking_var?Can I do:
y~te(x,cov1) + te(x,cov2) + te(x,cov3) + blocking_var
or should I create separate models for each interaction I'm trying to test? How would I account for possible influences on the various terms on each other?
Do I need to include a separate s(x) term? i.e.
y~ s(x)+ te(x,cov1) + te(x,cov2) + te(x,cov3) + blocking_varWhat is the difference between
te()andti()? That part I'm still not clear about.I visualized the partial effects with both
plot.gamandvis.gamand get vastly different plots depending on how I fit the model between plot.gam and vis.gam. I do not understand why the two ways to plot the model are so different, both among the two plotting functions as well as whether I uses(),te(), orti().
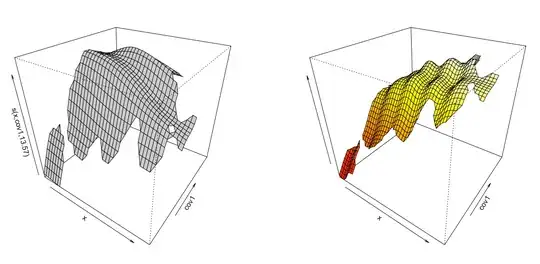
gam.1<-gam(y~ s(x) + s(x,cov1) + s(x,cov2) + s(x,cov3) + blocking_var, data=df, method = 'REML')
par(mfrow = c(1,2))
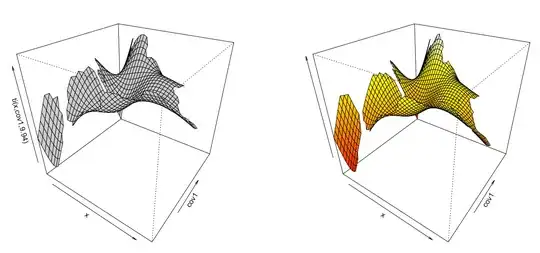
plot(gam.1, select=2, scheme=1, theta=35, phi=32,)
vis.gam(gam.1, view=c('x', 'cov1'), n.grid=50, theta=35, phi=32, zlab="", too.far=0.1)
gam.2<-gam(y~ s(x) + te(x,cov1) + te(x,cov2) + te(x,cov3) + blocking_var, data=df, method = 'REML')
plot(gam.2, select=2, scheme=1, theta=35, phi=32,)
vis.gam(gam.2, view=c('x', 'cov1'), n.grid=50, theta=35, phi=32, zlab="", too.far=0.1)
gam.3<-gam(y~ te(x) + te(x,cov1) + te(x,cov2) + te(x,cov3) + blocking_var, data=df, method = 'REML')
plot(gam.3, select=2, scheme=1, theta=35, phi=32,)
vis.gam(gam.3, view=c('x', 'cov1'), n.grid=50, theta=35, phi=32, zlab="", too.far=0.1)
gam.4<-gam(y~ te(x,cov1) + te(x,cov2) + te(x,cov3) + blocking_var, data=df, method = 'REML')
plot(gam.4, select=1, scheme=1, theta=35, phi=32,)
vis.gam(gam.4, view=c('x', 'cov1'), n.grid=50, theta=35, phi=32, zlab="", too.far=0.1)
gam.5<-gam(y~ ti(x,cov1) + ti(x,cov2) + ti(x,cov3) + blocking_var, data=df, method = 'REML')
plot(gam.5, select=1, scheme=1, theta=35, phi=32,)
vis.gam(gam.5, view=c('x', 'cov1'), n.grid=50, theta=35, phi=32, zlab="", too.far=0.1)
This obviously also has impact on subsequent maximum likelihood tests and whether or not terms/interactions are significant.
Thank you.
Edit: Following the really helpful reply by Isabella, I wanted to update my post with some more information regarding my specific data/modeling needs.
We know that:
y~s(x)
y~s(cov1)
x~s(cov1)
Therefore, x and cov1 are highly collinear. For our specific question, we're only interested in the relationsship y~x. This relationship is actually almost linear (it's quadratic), but for modelling purposes we still chose a GAM due to probable nonlinear interactions. We are intersted in how that simple y~xrelationsship changes qualitatively due to the impact of the other factors.
Cov1 is a temporal variable.
Cov2 and Cov3 are derivates of x, i.e they're not independent of x. They are proportions of specific groups of x per x (e.g. %females of counts x). There may be some slight collinearity, but if so it's not strong. Cov2 and Cov3 are not practical/causal predictors of y, therefore I should not include them as separate fixed effects, as far as I understand it.
We are not trying to find the best possible model describing our data. We're trying to describe qualitative modifications of covariates on our simple regression that is our primary interest.
Given those information, do I conclude correctly that the best approach is to use all te() terms?
(a) y~te(x,cov1) + te(x,cov2) + te(x, cov3) + blocking_var
or should I do something like
(b) y~te(x,cov1) + ti(x,cov2) + ti(x, cov3) + blocking_var ?
If b, is it sufficient to include te(x,cov1) instead of an additional ti(x) when using ti() for the other two covariates? As mentioned above, cov2 and cov3 should not be included as fixed effects; at least I wouldn't understand the practical (in my case biological) logic behind it.
Edit2: Adding another question.
- How do I interpret maximum likelihood ratio tests with
te()? With GLM with interactions, when I run a Type II anova, I will get p values for the fixed main effects and the interaction. A significant interaction means my significant main effect 1 isn't independent of value/factor of covariate 1.
With te(), I don't have separate main effects. What does a significant te() mean? How do I interpet this? I would assume I would interpet results from y~ti(x) + ti(x,cov1) the same as in a GLM. But how about y~te(x,cov1). Does it only mean x isn't independent of cov1, but what does it tell me about my response? How will I know which are my main predictors of y?