I have a data set with several hundred variables and some thousand records. I'm reviewing the different ways for running a Principal Component Analysis and choosing the principal components.
First I used the function prcomp() to get the advantage of using SVD. The first component explains more than 50% of the variance and the second a % more. The remaining components are very small.
This is the figure I got:
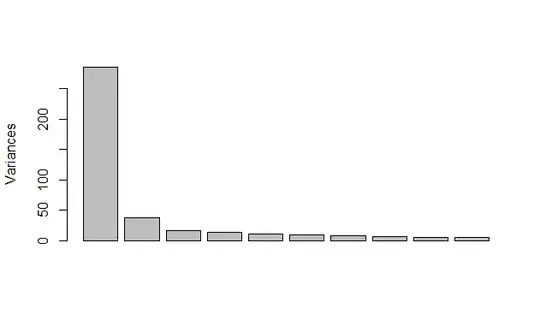
Then, by reading some posts here in Cross Validated I found that a recommended method for choosing the principal components was the so called "parallel analysis" and 2 libraries were recommended including the psych.
So I decided to try that method. This is the code I used:
> fa.parallel(my.dataframe,
fa="PC",
n.iter=100,
how.legend=FALSE,
main="My Plot")
I received the following results in the command prompt:
Loading required package: MASS
In smc, the correlation matrix was not invertible, smc's returned as 1s
In smc, the correlation matrix was not invertible, smc's returned as 1s
The determinant of the smoothed correlation was zero.
This means the objective function is not defined.
Chi square is based upon observed residuals.
The determinant of the smoothed correlation was zero.
This means the objective function is not defined for the null model either.
The Chi square is thus based upon observed correlations.
In factor.stats, the correlation matrix is singular, an approximation is used
In factor.scores, the correlation matrix is singular, an approximation is used
I was unable to calculate the factor score weights, factor loadings used instead
Parallel analysis suggests that the number of
factors = 54 and the number of components = 44
Warning messages:
1: In cor.smooth(R) : Matrix was not positive definite, smoothing was done
2: In cor.smooth(R) : Matrix was not positive definite, smoothing was done
3: In cor.smooth(r) : Matrix was not positive definite, smoothing was done
4: In factor.stats(r, loadings, Phi, n.obs = n.obs, np.obs = np.obs, :
In factor.stats, the correlation matrix is singular, and we could not calculate
the beta weights for factor score estimates
5: In cor.smooth(r) : Matrix was not positive definite, smoothing was done
And this is the resulting figure:
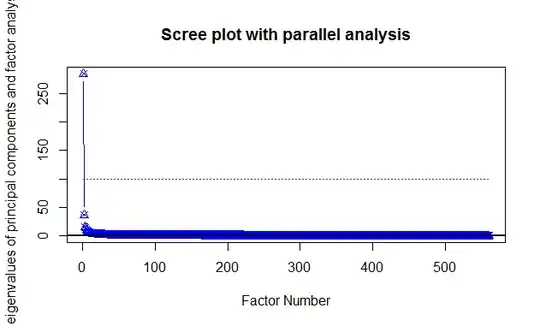
None of the PCA figures I had reviewed look like this, and I'm totally lost at analyzing its significance. I guess I should choose the first 44 principal components.
I'd really appreciate if somebody could explain this to me a little bit.