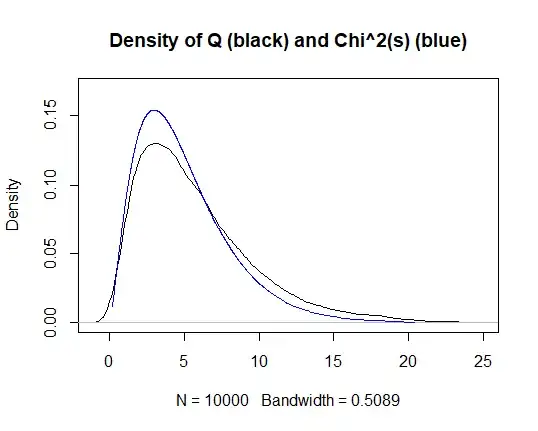
An $\text{MA}(h-1)$ process implies the autocorrelations beyond lag $h-1$ are zero. Let us adapt the Ljung-Box (LB) test to ignore the first $h-1$ autocorrelations and test a null hypothesis that autocorrelations at lags between $h$ and $h-1+s$ are jointly zero: $$ H_0\colon \ \rho_{h}=\rho_{h+1}=\dots=\rho_{h-1+s}=0. $$ Since the true autocorrelations of an $\text{MA}(h-1)$ process are zero, the test statistic should have a $\chi^2(s)$ distribution when applied on such a process. I have implemented this idea an carried out a simulation in R as described below, but I am not getting the expected result.
Question: What is my mistake?
Implementation in terms of a software function that calculates the LB statistic (such as stats::Box.test in R) is straightforward:
- Calculate the LB test statistic for lags up to $h-1+s$, $$\quad Q(h-1+s)=n(n+2)\sum_{k=1}^{h-1+s}\frac{\hat\rho_k^2}{n-k}.$$
- Calculate the LB test statistic for lags up to $h-1$, $$Q(h-1)=n(n+2)\sum_{k=1}^{h-1}\frac{\hat\rho_k^2}{n-k}.$$
- Subtract $Q(h-1)$ from $Q(h-1+s)$ to yield $$\quad Q(h,h-1+s)=\sum_{k=h}^{h-1+s}\frac{\hat\rho_k^2}{n-k}$$ and use it.
An R simulation below illustrates that something is not right. The test statistic does not have its theoretical distribution.
R code:
T=1e4 # length of time series
m=1e4 # number of simulations
h=4
s=5
Qlong=Qshort=rep(NA,m)
for(i in 1:m){
set.seed(i)
x=arima.sim(model=list(ma=c(0.6,-0.4,0.2)),n=T)
Qlong [i]=stats::Box.test(x=x,lag=h-1+s,type="Ljung-Box")$statistic
Qshort[i]=stats::Box.test(x=x,lag=h-1 ,type="Ljung-Box")$statistic
}
Q=Qlong-Qshort
mean(Q) # should be s
var (Q) # should be 2*s
# Plot kernel density of Q versus theoretical density of Chi^2(s)
plot(density(Q),xlim=c(-1,25),ylim=c(0,0.17),main="Density of Q (black) and Chi^2(s) (blue)")
q=qchisq(p=seq(from=0.001,to=0.999,by=0.001),df=s)
lines(y=dchisq(x=q,df=s),x=q,col="blue")
# Kolmogorov-Smirnov test
ks.test(Q,"pchisq",s)
Related questions: