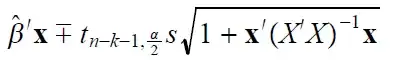The issue is that the units of the $X$-variables are not constant, so standardizing them makes them the same (at least in the sense of all being standard deviations—whether that really makes them the same is a bit of a philosophical issue).
You are discussing this in terms of variable importance, but the topic has been discussed extensively in the area of penalized estimation methods (i.e., ridge, LASSO, and elastic net regression). Hastie and Tibshirani have argued that you should standardize your dummy variables as well. If your factor is perfectly 50-50, this will output essentially $-1$ and $1$ as the new values; if it's unbalanced, it will shift towards $-\infty$ and $0$ or $0$ and $\infty$, depending on whether your (current) $0$'s or $1$'s are more prevalent, how imballanced they are, and how many data you have. This gets trickier if you have multi-category categorical variables. It may help you to read these threads:
A different way to visualize variable importance with a mix of categorical and continuous variables is to get a variable's chi-squared statistic from its likelihood ratio test and divide that by its degrees of freedom.
lrts = drop1(glm(Solea_solea ~ ., family="binomial", data=Solea), test="LRT")
## in this case, all variables have 1 df, so the division is a waste of time,
## but in other contexts, you could do:
lrts$importance = with(lrts, LRT/Df)
lrts
# Df Deviance AIC LRT Pr(>Chi) importance
# <none> 51.830 77.830
# Sample 1 53.762 77.762 1.9314 0.16461 1.9314
# season 1 52.101 76.101 0.2711 0.60259 0.2711
# month 1 53.341 77.341 1.5107 0.21903 1.5107
# Area 1 58.696 82.696 6.8660 0.00879 6.8660
# depth 1 51.957 75.957 0.1273 0.72125 0.1273
# temperature 1 51.922 75.922 0.0918 0.76190 0.0918
# salinity 1 55.457 79.457 3.6269 0.05685 3.6269
# transparency 1 52.125 76.125 0.2953 0.58688 0.2953
# gravel 1 51.834 75.834 0.0039 0.95020 0.0039
# large_sand 1 51.834 75.834 0.0041 0.94922 0.0041
# med_fine_sand 1 51.834 75.834 0.0043 0.94800 0.0043
# mud 1 51.834 75.834 0.0041 0.94888 0.0041