I am trying to use hurdle gamma model for one of my use cases, to handle a zero-inflated scenario. I have a very simple code creating dummy data with quite a few zeros.
# Dataset prep
non_zero <- rbinom(1000, 1, 0.1)
g_vals <- rgamma(n = 1000, shape = 2, scale = 2)
dat <- data.frame(x = non_zero * g_vals)
The model is written as
hum <- brm(bf(x ~ 1, hu ~ 1), data = dat, family = hurdle_gamma)
I would like to understand the results and the associated parameters.
A plot of the predicted results from the model using
tibble(x=1) %>% add_fitted_draws(hum) %>% ggplot(aes(x = .value)) + geom_density()
is as follows
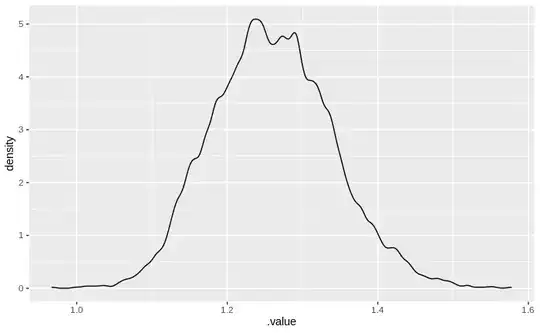 The posterior summary is:
The posterior summary is:
Estimate Est.Error Q2.5 Q97.5
b_Intercept 1.468677 0.1037202 1.271352 1.681500
b_hu_Intercept 2.081498 0.1474433 1.802057 2.372279
shape 1.757053 0.3114681 1.203522 2.418776
lp__ -315.947968 1.2657819 -319.090956 -314.508678
I don't see any divergences in the model fit. Also there is pretty good mixing of the chains for the parameters. Given the whole reason for me to consider the hurdle model was to see a model predict zeros in abundance, I am unable to understand the predictions. Shouldn't I see a lot of zeros?
It would be great if someone can throw light on the share a simple test case on using the hurdle model. I am unable to find a nice write up of modeling using hurdle_gamma using brms.
- Operating System: Ubuntu 18.04
- brms Version:2.13