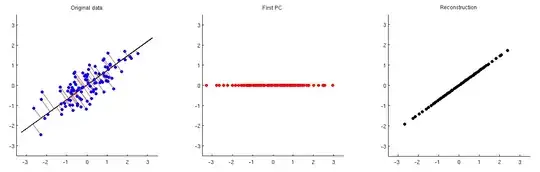
I try to replicate this graph, which appears in this question with no luck.
I post it here as I believe that this question is more about PCA understanding rather than coding/programming itself.
# Code based on a question from: https://stats.stackexchange.com/questions/229092/how-to-reverse-pca-and-reconstruct-original-variables-from-several-principal-com
library(ggplot2)
X = iris[,1:4]
mu = colMeans(X)
Xpca = prcomp(X)
nComp = 1
Xhat = Xpca$x[,1:nComp] %*% t(Xpca$rotation[,1:nComp])
Xhat = scale(Xhat, center = -mu, scale = FALSE)
load <- Xpca$rotation
slope <- load[2, ]/load[1, ]
intcpt <- mu[2] - (slope * mu[1])
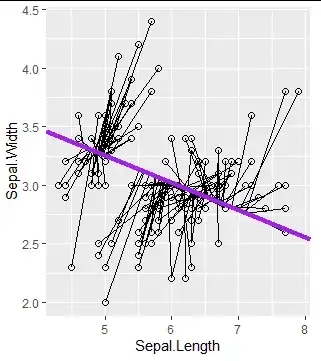
gg = ggplot(iris[,1:2], aes(x = Sepal.Length, y = Sepal.Width)) +
geom_point(size = 2, shape = 21)
#Project first PCA back to the feature space
gg+geom_segment(xend = Xhat[,1], yend = Xhat[,2])+
geom_abline(intercept = intcpt[1], slope = slope[1], col = "purple", lwd = 1.5)
I expected to see orthogonal lines between the slope (ratio) between the two feature based on the first PC, but this is not the case.
As an illustration, I plot the expected segments. My understanig is that the first pc will project this data points to a single sub-space (i.e the purple line).
#calculate the perpendicular segment manually
perp.segment.coord <- function(x0, y0, intcpt,slope){
# Code based on question from: https://stackoverflow.com/a/30399576/786542
#finds endpoint for a perpendicular segment from the point (x0,y0) to the line
# defined by lm.mod as y=a+b*x
a <- intcpt #intercept
b <- slope #slope
x1 <- (x0+b*y0-a*b)/(1+b^2)
y1 <- a + b*x1
list(x0=x0, y0=y0, x1=x1, y1=y1)
}
ss <- perp.segment.coord(iris[,1], iris[,2],intcpt=intcpt[1],slope=slope[1] )
gg +geom_segment(xend = ss$x1, yend = ss$y1)+
geom_abline(intercept = intcpt[1], slope = slope[1], col = "purple", lwd = 1.5)