I'm trying to model the data for covid-19 using SIR model in R. I followed the answer of the question, and the blog. I'm using the suggested code, However, the data does not converging. Any suggestion what I'm missing.
# required libraries
library(deSolve)
library(shape) # for plotting arrows
library(progress) # for drawing the progress bar
####################################
##
## The basic model (which does not result in the exact solution)
## Adapted from the previous blogpost but with small adaptations
##
####################################
# the data infected represents cumalative sum (cumsum (infected)- (cumsum(recovered)+cumsum(death) ) )
IpRpD <- c(5,11,26,43,45,45,46,56,56,56,57,57,60,63,63,67,67,75,95,
97,103,111,118,127,130,137,149,158,159,152,152,159,168,
171,188,194,216,237,261,335,385,456,561,637,743,798,869,
1020,1091,1148,1176,1196,1296,1395,1465,1603,1619,1657,1792,
1887,1986,2217,2249,2254,2241,2327,2459,2745,2883,3169,3291,
3732,4028,4142,4695,4952,5901,6314,7101,7683,8436,9124,9852,
10645,11234,11962,12559)
# only recovered not cummalative sum
R <- c(310,320,204,342,246,250,203,189,188,162,194,178,107,156,85,162,187,85,171,73,101,63,150,
213,164,206,150,43,115,55,31,45,62,25,22,33,19,30,26,8,9,10,12,0,6,2,4,6,11,1,1,7,1,5,3,
7,8,6,4,9,0,3,5,4,3,3,0,3,2,2,0,0,3,0,1,0,0,0,1,0,0,0,0,0,0,0,0,0,0)
# Only death NOT cuumalative
D <- c(9,5,3,3,6,5,11,8,6,7,10,7,9,2,3,2,2,0,2,5,3,4,2,1,1,2,1,4,1,1,2,2,2,1,1,2,0,0,1,1,0,0,0,0,
0,0,0,0,1,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0)
Infected <-IpRpD
Day <- 1:(length(Infected))
N <- 4921638 #population
# ODE equation used for fitting
#
# I have removed the R(t) in comparison
# to the function used in the odler blogpost
# because we are not gonna use that value
# also we have anyway: R(t) = N(0) - N(t) - I(t)
SIR <- function(time, state, parameters) {
par <- as.list(c(state, parameters))
with(par, {
dS <- -beta/N * I * S
dI <- beta/N * I * S - gamma * I
list(c(dS, dI))
})
}
#
# cost function to be optimized in the fitting
#
RSS <- function(parameters) {
names(parameters) <- c("beta", "gamma")
out <- ode(y = init, times = Day, func = SIR, parms = parameters)
fitInfected <- out[,3]
# fitInfected <- N-out[,2] # this would be a better comparison since the data is not the number of Infectious people
sum((Infected - fitInfected)^2)
}
# starting condition
init <- c(S = N-Infected[1], I = Infected[1])
# init <- c(S = N-Infected[1], I = Infected[1]-R[1]-D[1]) use this starting condition when applying the different line in the RSS function
# performing the fit
Opt <- optim(c(0.5, 0.5), RSS, method = "L-BFGS-B", lower = c(0, 0), upper = c(1, 1)) # optimize with some sensible conditions
Opt$message
Opt_par <- setNames(Opt$par, c("beta", "gamma"))
Opt_par
## beta gamma
# plotting the result
t <- 1:120 # time in days
fit <- data.frame(ode(y = init, times = t, func = SIR, parms = Opt_par))
plot(Day,Infected, xlim = range(Day), ylim = range(Infected) )
lines(t,fit[,3])
###########################
##
## Alternative model which provides a better fit
##
############################
# We transform the equations and instead of
# parameters beta and gamma
# we use parameters
#
# K = beta-gamma
# R0 = beta/gamma
#
# or
#
# beta = K * R0/(R0-1)
# gamma = K * 1/(R0-1)
#
# then the equations become
#
# dS = I * K * (-S/N * R0)/(R0-1)
# dI = I * K * ( S/N * R0 - 1)/(R0-1)
# note in the beginning, S/N = 1
# then in the start you get this approximate exponential growth
# dI = I * K * (1)
SIR2 <- function(time, state, parameters) {
par <- as.list(c(state, parameters))
with(par, {
dS <- I * K * (-S/N * R0/(R0-1))
dI <- I * K * ( S/N * R0/(R0-1) - 1/(R0-1))
list(c(dS, dI))
})
}
RSS2 <- function(parameters) {
names(parameters) <- c("K", "R0")
out <- ode(y = init, times = Day, func = SIR2, parms = parameters)
fitInfected <- out[,3]
#fitInfected <- N-out[,2]
sum((Infected - fitInfected)^2)
}
### Two functions RSS to do the optimization in a nested way
###
### This nesting requires a lot more computational power
### However, it makes that we have to worry less about the different scale
### of the parameters
Infected_MC <- Infected
SIRMC2 <- function(R0,K) {
parameters <- c(K=K, R0=R0)
out <- ode(y = init, times = Day, func = SIR2, parms = parameters)
fitInfected <- out[,3]
#fitInfected <- N-out[,2]
RSS <- sum((Infected_MC - fitInfected)^2)
return(RSS)
}
SIRMC <- function(K) {
optimize(SIRMC2, lower=1,upper=10^5,K=K, tol = .Machine$double.eps)$objective
}
# wrapper to optimize and return estimated values
getOptim <- function() {
opt1 <- optimize(SIRMC,lower=0,upper=1, tol = .Machine$double.eps)
opt2 <- optimize(SIRMC2, lower=1,upper=10^5,K=opt1$minimum, tol = .Machine$double.eps)
return(list(RSS=opt2$objective,K=opt1$minimum,R0=opt2$minimum))
}
# starting condition
#init <- c(S = N-Infected[1], I = Infected[1]-R[1]-D[1])
init <- c(S = N-Infected[1], I = Infected[1])
# performing the fit
# starting K=0.3, R0 = 2
Opt2 <- optim(c(0.3, 2), RSS2, method = "L-BFGS-B",
hessian = TRUE, control = list(parscale = c(10^0,10^0), factr = 1))
Opt2
Opt3 <- getOptim()
Opt3
Opt_par2 <- setNames(Opt2$par, c("K", "R0"))
Opt_par3 <- setNames(Opt3[2:3], c("K", "R0"))
# plotting the result
t <- seq(1,120,1) # time in days
fit1 <- data.frame(ode(y = init, times = t, func = SIR , parms = Opt_par))
fit2 <- data.frame(ode(y = init, times = t, func = SIR2, parms = Opt_par2))
fit3 <- data.frame(ode(y = init, times = t, func = SIR2, parms = Opt_par3))
plot(Day,Infected, xlim = range(Day), ylim = range(Infected),
log = "", xaxt = "n",
main = "Infected(including Recovered and Death)", xlab = "Day", ylab = "number infected")
lines(t, fit3[,3], col = 1)
lines(t, fit2[,3], col = 4, lty = 2)
lines(t, fit1[,3], col = 2, lty = 3)
axis(1, at = 1:90, labels = rep("",90), tck = -0.01)
# these labels are not valid for the current date
axis(1, at = c(1,8,15,22), labels = c("Jan 16", "Jan 23", "Jan 30", "Feb 6"))
text(t[183]+2,fit1[183,3]+1800,"old optim fit",pos=4, col=2)
text(t[183]+2,fit1[183,3],expression(R[0] == 2.07),pos=4, col=2)
text(t[183]+2,fit1[183,3]-1400,expression(RSS == 74.3 %*% 10^6),pos=4, col=2)
text(t[220]+3,fit2[220,3]+3200,"new optim fit",pos=3, col=4)
text(t[220]+3,fit2[220,3]+1400,expression(R[0] == 1.0054626),pos=3, col=4)
text(t[220]+3,fit2[220,3],expression(RSS == 6.5 %*% 10^6),pos=3, col=4)
text(t[240]-3,fit3[240,3],"nested algorithm",pos=1, col=1)
text(t[240]-3,fit3[240,3]+700-2500,expression(R[0] == 1.005332),pos=1, col=1)
text(t[240]-3,fit3[240,3]-700-2500,expression(RSS == 5.9 %*% 10^6),pos=1, col=1)
x1 <- t[240]-3; x2 <- t[225];
y1 <- fit3[240,3]; y2 <- fit3[225,3]
Arrows(x1,y1,x1+(x2-x1)*0.65,y1+(y2-y1)*0.65, col = 1)
x1 <- t[220]+2; x2 <- t[227];
y1 <- fit2[220,3]; y2 <- fit2[227,3]
Arrows(x1,y1,x1+(x2-x1)*0.6,y1+(y2-y1)*0.6, col = 4)
x1 <- t[183]+2; x2 <- t[183];
y1 <- fit1[183,3]; y2 <- fit1[183,3]
Arrows(x1,y1,x1+(x2-x1)*0.6,y1+(y2-y1)*0.6, col = 2)
####################
##
## Graph with various values of R0
##
#######################
# starting condition
#init <- c(S = N-Infected[1], I = Infected[1]-R[1]-D[1])
init <- c(S = N-Infected[1], I = Infected[1])
Infected_MC <- Infected
SIRMC3 <- function(R0,K) {
parameters <- c(K=K, R0=R0)
out <- ode(y = init, times = Day, func = SIR2, parms = parameters)
fitInfected <- out[,3]
#fitInfected <- N-out[,2]
RSS <- sum((Infected_MC - fitInfected)^2)
return(RSS)
}
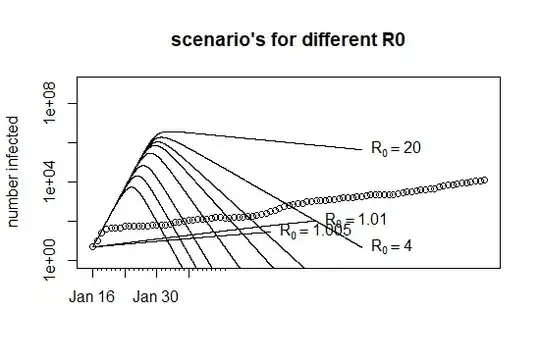
plot(Day,Infected, xlim = range(Day), ylim = c(1,10^9),
log = "y", xaxt = "n",
main = "scenario's for different R0", xlab = "", ylab = "number infected")
axis(1, at = 1:30, labels = rep("",30), tck = -0.01)
axis(1, at = c(1,8,15,22), labels = c("Jan 16", "Jan 23", "Jan 30", "Feb 6"))
for (i in 1:10) {
R0 <- c(1.005,1.01,1.05,1.1,1.2,1.5,2,2.5,4,20)[i]
K <- optimize(SIRMC3, lower=0,upper=1,R0=R0, tol = .Machine$double.eps)$minimum
parameters <- c(K,R0)
xd <- seq(1,60,0.01)
if (i == 1) {
xd <- seq(1,40,0.01)
}
if (i == 2) {
xd <- seq(1,50,0.01)
}
out <- ode(y = init, times = xd, func = SIR2, parms = parameters)
lines(xd,out[,3])
text(tail(xd,1),tail(out[,3],1),bquote(R[0] == .(R0)), pos =4)
}
