If I understand you correctly, you want to know whether you can reliably classify your participants based on their early weight loss. So it's basically a question of classification. I recon you also have a training data set, with measurements $W0$, $W1$, and $W2$. Let's assume your data look like this:
or, relative to the baseline:
You could, as you suggested, declare some arbitrary boundary (0.02) for the effective weight loss and label your data accordingly:
The simplest thing to do now, if you just want to know whether the early weight loss is indicative for the later weight loss, would be to run a t-test on the x-axis (the early weight loss) of the two groups. In Python:
import numpy as np
import matplotlib.pyplot as plt
mu0 = 90
sigma0 = 15
sigma1 = .2
sigma2 = 1
Na = 20
Nb = 200
effect1 = .99
effect2 = .975
rng = np.random.RandomState(42)
w0 = rng.normal(mu0, sigma0, Na+Nb)
w1 = np.concatenate([
(effect1 * w0)[:Na ] + rng.normal(0, sigma1, Na),
( w0)[ Na:] + rng.normal(0, sigma1, Nb)
])
w2 = np.concatenate([
(effect2 * w0)[:Na ] + rng.normal(0, sigma2, Na),
( w0)[ Na:] + rng.normal(0, sigma2, Nb)
])
r1 = (w0-w1)/w0
r2 = (w0-w2)/w0
# t-test on the x-axis
from scipy import stats
tt = stats.ttest_ind(r1[r2<.02], r1[r2>.02])
print(tt)
produces:
Ttest_indResult(statistic=-11.912202742445448, pvalue=1.581946945815335e-25)
But you probably want to have a classification algorithm, so you'll need to train a model. A logistic regression will give you not only the probabilities of class membership, but also the p-values for the coefficients, thus answering your initial question:
# logistic regression
import statsmodels.api as sm
logit_model=sm.Logit(r2>.02, sm.add_constant(r1))
result=logit_model.fit()
print(result.summary())
gives you:
Logit Regression Results
==============================================================================
Dep. Variable: y No. Observations: 220
Model: Logit Df Residuals: 218
Method: MLE Df Model: 1
Date: Tue, 05 May 2020 Pseudo R-squ.: 0.4436
Time: 21:25:18 Log-Likelihood: -42.182
converged: True LL-Null: -75.814
Covariance Type: nonrobust LLR p-value: 2.373e-16
==============================================================================
coef std err z P>|z| [0.025 0.975]
------------------------------------------------------------------------------
const -3.4028 0.405 -8.393 0.000 -4.197 -2.608
x1 472.5083 77.561 6.092 0.000 320.491 624.526
==============================================================================
If you want to make it less arbitrary and more sophisticated, you can, instead of arbitrary labeling, use clustering (e.g. K-means) on the two-dimensional data and use cluster labels for training your classifier:
from sklearn.cluster import KMeans
# important: need to scale the axes appropriately!
X_SCALE = (np.max(r2) - np.min(r2)) / (np.max(r1) - np.min(r1))
w = np.concatenate([X_SCALE*r1.reshape([-1, 1]), r2.reshape([-1, 1])], axis=1)
k2 = KMeans(n_clusters=2).fit(w)
fig, ax = plt.subplots()
ax.plot(r1[k2.labels_==0], r2[k2.labels_==0], '.', c='orangered')
ax.plot(r1[k2.labels_==1], r2[k2.labels_==1], '.', c='green')
ax.set(xlabel='(W0 - W1) / W0', ylabel='(W0 - W2) / W0', title='Relative Weight Loss, Clustered')
ax.grid()
ax.legend(["Not working", "Working"])
plt.show()

Logistic regression on data labeled this way is slightly different:
Logit Regression Results
==============================================================================
Dep. Variable: y No. Observations: 220
Model: Logit Df Residuals: 218
Method: MLE Df Model: 1
Date: Tue, 05 May 2020 Pseudo R-squ.: 0.8536
Time: 21:30:47 Log-Likelihood: -10.787
converged: True LL-Null: -73.691
Covariance Type: nonrobust LLR p-value: 3.389e-29
==============================================================================
coef std err z P>|z| [0.025 0.975]
------------------------------------------------------------------------------
const -7.7915 2.166 -3.597 0.000 -12.037 -3.546
x1 1732.4376 614.892 2.817 0.005 527.271 2937.604
==============================================================================
The following graph shows the logistic regression for arbitrary (manual cutoff) labeling and for K-means labeling:
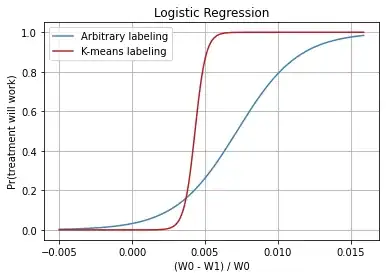
Update:
Spurious correlation ("mathematical coupling") is not an issue here. Observe a set where the long-term weight loss is not related to the early weight loss:
rng = np.random.RandomState(42)
w0 = rng.normal(mu0, sigma0, Na+Nb)
w1 = np.concatenate([
(effect1 * w0)[:Na ] + rng.normal(0, sigma1, Na),
( w0)[ Na:] + rng.normal(0, sigma1, Nb)
])
w2 = w0 + rng.normal(0, 2*sigma1, Na+Nb)
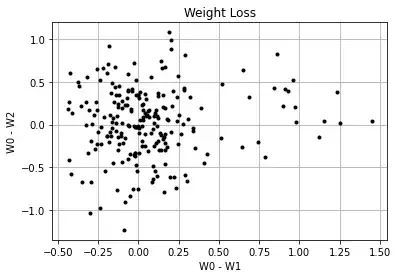
You can define a "meaningful" weight loss to be as low as 0.5%, but it will be equally distributed over the two groups, with and without early weight loss:

The mean relative early weight losses won't differ between the groups:
stats.ttest_ind(r1[r2<.005], r1[r2>.005])
Ttest_indResult(statistic=-0.17757452954060468, pvalue=0.8592220151288611)
and you'll get the same p-value in logistic regression.
If you do K-means, it will find two clusters, but along the x-axis (early weight loss). You need to check that the long-term losses differ in the two clusters.







